| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,415,979 – 25,416,071 |
| Length | 92 |
| Max. P | 0.757415 |

| Location | 25,415,979 – 25,416,071 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -14.61 |
| Energy contribution | -15.83 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
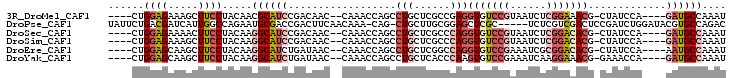
>3R_DroMel_CAF1 25415979 92 + 27905053 AUUUGGCAUC----UGGAUAG-CGUUUCCGAGAUUACGGACACCUCGGCGAGCAGGCUGGUUUG--GUUGUCGGAUGCGUUGUAGGAAGCUUUCUCCAG---- ....(((.((----(..((((-(((.(((((.(...(((((....((((......)))))))))--..).))))).))))))).))).)))........---- ( -30.10) >DroPse_CAF1 110456 96 + 1 GUCUGGCACGUAUCCAGAUCGGAGUCGACGAGA-----GCGAGCUCCGCAAGCAG-CUG-UUUGUUGAAGUCGGUCGCAUUCUGGCCAAUGAUCGUCAGAAUA .((((((.........((((((..((((((((.-----...((((.(....).))-)).-))))))))..))))))((......))........))))))... ( -32.90) >DroSec_CAF1 99727 92 + 1 AUUUGGCAUC----UGGAUAG-CGUGUCCGAGAUUACGGACACCUGGGCGAGCAGGCUGGUUUG--GUUGUCGGAUGCCUUGUAGGAAGUUUUCUCCAG---- ((..((((((----((.((((-((((((((......))))))).(..((.((....)).))..)--)))))))))))))..)).(((.......)))..---- ( -36.30) >DroSim_CAF1 100234 92 + 1 AUUUGGCAUC----UGGAUAG-CGUGUCCGAGAUUACGGACACCUGGGCGAGCAGGCUGGUUUG--GUUGUCGGAUGCCUUGUAGGAAGCUUUCUCCAG---- ((..((((((----((.((((-((((((((......))))))).(..((.((....)).))..)--)))))))))))))..)).(((.......)))..---- ( -36.30) >DroEre_CAF1 106065 92 + 1 AUUUGGCAUU----UGGAUAG-CGUGUCCGCGAUUUCGGACACCUGGCCGAGCAGGCUGGUUUG--GUUAUCAGAUGCCUUGUAGGAAGCUUGCUCCAG---- ((..((((((----((.((((-((((((((......)))))))(..(((.....)))..)....--)))))))))))))..)).(((.(....))))..---- ( -38.30) >DroYak_CAF1 107828 92 + 1 AUUUGGCAUC----UGGUUUC-CGUUUCCUUGAUUUCGGACACUUGGGUGAGCAGGCUGGUUUG--GUUAUCAGAUGCCUUGUAGGAAGCUUGCUCCAG---- ((..((((((----((((..(-((..((((((...(((..(....)..))).))))..))..))--)..))))))))))..)).(((.(....))))..---- ( -29.00) >consensus AUUUGGCAUC____UGGAUAG_CGUGUCCGAGAUUACGGACACCUGGGCGAGCAGGCUGGUUUG__GUUGUCGGAUGCCUUGUAGGAAGCUUUCUCCAG____ ....((((((.............(((((((......))))))).....(((((......))))).........)))))).....(((.......)))...... (-14.61 = -15.83 + 1.23)



| Location | 25,415,979 – 25,416,071 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -13.94 |
| Energy contribution | -15.75 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25415979 92 - 27905053 ----CUGGAGAAAGCUUCCUACAACGCAUCCGACAAC--CAAACCAGCCUGCUCGCCGAGGUGUCCGUAAUCUCGGAAACG-CUAUCCA----GAUGCCAAAU ----(((((...(((..........(((..(......--.......)..)))...(((((((.......)))))))....)-)).))))----)......... ( -21.12) >DroPse_CAF1 110456 96 - 1 UAUUCUGACGAUCAUUGGCCAGAAUGCGACCGACUUCAACAAA-CAG-CUGCUUGCGGAGCUCGC-----UCUCGUCGACUCCGAUCUGGAUACGUGCCAGAC ...((((.((........(((((.((.((.((((.........-..(-(.....))((((....)-----))).))))..)))).))))).....)).)))). ( -22.42) >DroSec_CAF1 99727 92 - 1 ----CUGGAGAAAACUUCCUACAAGGCAUCCGACAAC--CAAACCAGCCUGCUCGCCCAGGUGUCCGUAAUCUCGGACACG-CUAUCCA----GAUGCCAAAU ----..((((.....)))).....((((((.(.....--.......(((((......)))))(((((......)))))...-.....).----)))))).... ( -26.90) >DroSim_CAF1 100234 92 - 1 ----CUGGAGAAAGCUUCCUACAAGGCAUCCGACAAC--CAAACCAGCCUGCUCGCCCAGGUGUCCGUAAUCUCGGACACG-CUAUCCA----GAUGCCAAAU ----..((((.....)))).....((((((.(.....--.......(((((......)))))(((((......)))))...-.....).----)))))).... ( -26.90) >DroEre_CAF1 106065 92 - 1 ----CUGGAGCAAGCUUCCUACAAGGCAUCUGAUAAC--CAAACCAGCCUGCUCGGCCAGGUGUCCGAAAUCGCGGACACG-CUAUCCA----AAUGCCAAAU ----..((((....))))......(((((.((.....--.......(((.....))).(((((((((......))))))).-))...))----.))))).... ( -27.60) >DroYak_CAF1 107828 92 - 1 ----CUGGAGCAAGCUUCCUACAAGGCAUCUGAUAAC--CAAACCAGCCUGCUCACCCAAGUGUCCGAAAUCAAGGAAACG-GAAACCA----GAUGCCAAAU ----..((((....))))......((((((((.....--.....................((.(((........))).))(-....)))----)))))).... ( -23.30) >consensus ____CUGGAGAAAGCUUCCUACAAGGCAUCCGACAAC__CAAACCAGCCUGCUCGCCCAGGUGUCCGUAAUCUCGGACACG_CUAUCCA____GAUGCCAAAU ......((((.....)))).....((((((..................(((......)))(((((((......))))))).............)))))).... (-13.94 = -15.75 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:31 2006