
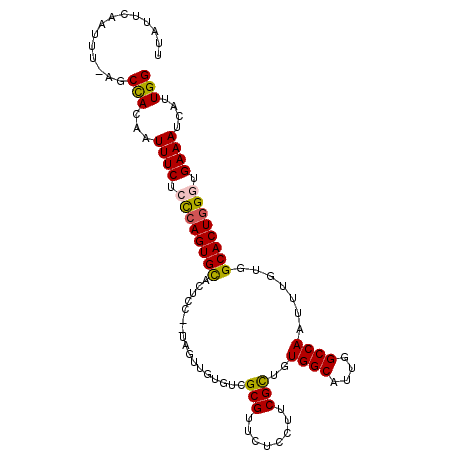
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,410,269 – 25,410,373 |
| Length | 104 |
| Max. P | 0.614793 |

| Location | 25,410,269 – 25,410,373 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.70 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -26.00 |
| Energy contribution | -25.83 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25410269 104 + 27905053 UUAUUCAACUU-GGCUACACUUUCUCCCAGUGCACUCCUCUAGUUGUGUCGCGUUCUCCUUCGCUGUGGCAUUGGCCAAUUUGUGGCACUGGGUGAAAUCAUUGG ...........-........((((.((((((((((...............(((........)))..((((....))))....)).)))))))).))))....... ( -28.60) >DroSec_CAF1 94201 102 + 1 UUAUUAAAUUU-AGCUACAAUUUCUCCCAGUGCACUCC--UAGUUGUGUCGCGUUCUCCUUCGCUGUGGCAUUGGCCAAUUUGUGGCACUGGGUGAAAUCAUUGG ...........-.......(((((.((((((((((...--..........(((........)))..((((....))))....)).)))))))).)))))...... ( -29.40) >DroSim_CAF1 94635 102 + 1 CUAUUCAAUUU-AGCUACAAUUUCUCCCAGUGCACUCC--UAGUUGUGUCGCGUUCUCCUUCGCUGUGGCAUUGGCCAAUUUGUGGCACUGGGUGAAAUCAUUGG ...........-.......(((((.((((((((((...--..........(((........)))..((((....))))....)).)))))))).)))))...... ( -29.40) >DroEre_CAF1 98132 102 + 1 UUGAUCAUUUU-AGCCACCAUUUCUGCCAGUGCAGUCC--UAGUAGUGCCGCGGUCUCCUUCGCUGUGGCAUUGGCCAAUUUGUGGCACUGGGUGAAAUCAUUGG (..((.(((((-(..........((((....)))).((--(((((((((((((((.......))))))))))).((((.....)))))))))))))))).))..) ( -34.20) >DroYak_CAF1 101985 102 + 1 UUUUUAAUUGU-AGCCAUAUUUUCUGCCAGUGCAUUAC--UAGUAGUGUCGCGUUCUCCUUCGCGGUGGCAUUGGCCAAUUUGUGGCACUGGGUGAAAUCAUUGG .(((((...((-.((((((..((..((((((((((((.--...)))).(((((........)))))..)))))))).))..))))))))....)))))....... ( -30.80) >DroAna_CAF1 102181 99 + 1 ACAUUCGUUUAAAGCCA-AUUUUCUCUCAGUGUAC-----UGAUGGUAUCGCGUUCUCCUUCGUUGUGGCAUUGGCCAAUAUGUGGCACUAGGUGAAAUCAUUGG ..............(((-((((((.(..(((((.(-----..(((.(((.(((........))).))).)))..).((.....)))))))..).))))..))))) ( -22.40) >consensus UUAUUCAAUUU_AGCCACAAUUUCUCCCAGUGCACUCC__UAGUUGUGUCGCGUUCUCCUUCGCUGUGGCAUUGGCCAAUUUGUGGCACUGGGUGAAAUCAUUGG ..............(((...((((.((((((((.................(((........)))..((((....)))).......)))))))).))))....))) (-26.00 = -25.83 + -0.16)


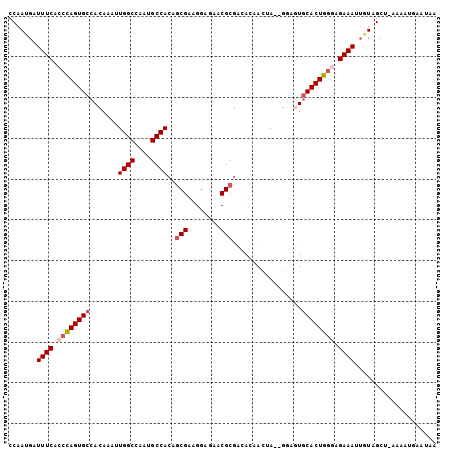
| Location | 25,410,269 – 25,410,373 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.70 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25410269 104 - 27905053 CCAAUGAUUUCACCCAGUGCCACAAAUUGGCCAAUGCCACAGCGAAGGAGAACGCGACACAACUAGAGGAGUGCACUGGGAGAAAGUGUAGCC-AAGUUGAAUAA .((((..((((.((((((((.((....((((....))))..(((........)))...............)))))))))).))))........-..))))..... ( -28.40) >DroSec_CAF1 94201 102 - 1 CCAAUGAUUUCACCCAGUGCCACAAAUUGGCCAAUGCCACAGCGAAGGAGAACGCGACACAACUA--GGAGUGCACUGGGAGAAAUUGUAGCU-AAAUUUAAUAA ...(..(((((.((((((((.((....((((....))))..(((........)))..........--...)))))))))).)))))..)....-........... ( -31.60) >DroSim_CAF1 94635 102 - 1 CCAAUGAUUUCACCCAGUGCCACAAAUUGGCCAAUGCCACAGCGAAGGAGAACGCGACACAACUA--GGAGUGCACUGGGAGAAAUUGUAGCU-AAAUUGAAUAG ...(..(((((.((((((((.((....((((....))))..(((........)))..........--...)))))))))).)))))..)....-........... ( -31.60) >DroEre_CAF1 98132 102 - 1 CCAAUGAUUUCACCCAGUGCCACAAAUUGGCCAAUGCCACAGCGAAGGAGACCGCGGCACUACUA--GGACUGCACUGGCAGAAAUGGUGGCU-AAAAUGAUCAA ....((((((((((.((((((......((((....))))..(((..(....))))))))))....--...((((....))))....)))))..-.....))))). ( -32.11) >DroYak_CAF1 101985 102 - 1 CCAAUGAUUUCACCCAGUGCCACAAAUUGGCCAAUGCCACCGCGAAGGAGAACGCGACACUACUA--GUAAUGCACUGGCAGAAAAUAUGGCU-ACAAUUAAAAA (((.((.((((..(((((((.......((((....)))).((((........)))).........--.....)))))))..)))).)))))..-........... ( -25.60) >DroAna_CAF1 102181 99 - 1 CCAAUGAUUUCACCUAGUGCCACAUAUUGGCCAAUGCCACAACGAAGGAGAACGCGAUACCAUCA-----GUACACUGAGAGAAAAU-UGGCUUUAAACGAAUGU (((((..((((.....(((........((((....))))...(....)....))).......(((-----(....))))))))..))-))).............. ( -17.60) >consensus CCAAUGAUUUCACCCAGUGCCACAAAUUGGCCAAUGCCACAGCGAAGGAGAACGCGACACAACUA__GGAGUGCACUGGGAGAAAUUGUAGCU_AAAAUGAAUAA .......((((.((((((((.......((((....))))..(((........))).................)))))))).)))).................... (-21.42 = -22.12 + 0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:30 2006