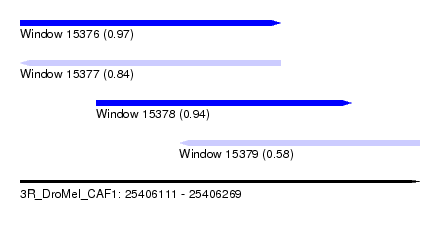
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,406,111 – 25,406,269 |
| Length | 158 |
| Max. P | 0.968188 |

| Location | 25,406,111 – 25,406,214 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -18.15 |
| Energy contribution | -19.32 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25406111 103 + 27905053 ----GGGGUAUGCGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACCGGCCAACGAAAUUGCAAAUAUACGAGUAACCACAU ----..(((.(((((.(((((((..((........)).....(((.((((((.......)))))).)))...)))))))....)))))..(((....)))))).... ( -31.50) >DroPse_CAF1 100139 98 + 1 ----GCUGGAGCUGGAGCUGAAGCCGGAGCCAAAGCGAUUCAUUGAAUGGCCAAAAACAGGCCAUCGAAACUGGCCAACAGAAUUGGGAAUAU-----UGCACACGC ----(((((..((((........))))..)))..(((((((.((((.(((((.......)))))))))..(((.....)))......))..))-----)))....)) ( -28.00) >DroEre_CAF1 94087 102 + 1 ----GGGGGCUUUGCAGUUGGCCACGAAGCCAAAGCCACUCAUUCAAUGGCCAA-AACGGGCCAUGGAAACUGGCCAACGAAAUUGCAAAUAUACGAGUGACCACAU ----((..((((((((((((((((....((....))......(((.((((((..-....)))))).)))..)))))))).....)))).......))))..)).... ( -33.51) >DroYak_CAF1 97613 107 + 1 ACAGGGGGGAUUUGGAGUUGGCCACGAAGCCAAAGCCUCUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACUGGCCAACGAAAUUGCAAAUAUACGAGUGACCACAU ...((((((.(((((..(((....)))..))))).))))((((((..((........))(((((.(....))))))...................)))))))).... ( -33.70) >DroAna_CAF1 98196 93 + 1 ---------AAACCGAGUUGGCCACGGAGCCAAAGCGAUUCAUUGAAUGGCCAAAAACAGGCCAUGGAAACUGGCCAACAAAAUUGCCAAUAUAUGG-----CACAU ---------.......((((((((.(...((....(((....))).((((((.......))))))))...))))))))).....(((((.....)))-----))... ( -31.90) >DroPer_CAF1 97420 98 + 1 ----GCUGGAGCUGGAGCUGAAGCCGGAGCCAAAGCGAUUCAUUGAAUGGCCAAAAACAGGCCAUCGAAACUGGCCAACAGAAUUGGGAAUAU-----UGCACACGC ----(((((..((((........))))..)))..(((((((.((((.(((((.......)))))))))..(((.....)))......))..))-----)))....)) ( -28.00) >consensus ____GGGGGAUCUGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACAGGCCAUGGAAACUGGCCAACAAAAUUGCAAAUAUACGA_UGACCACAU ................(((((((.....((....))......((((.(((((.......)))))))))....)))))))............................ (-18.15 = -19.32 + 1.17)

| Location | 25,406,111 – 25,406,214 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -19.68 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.844957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25406111 103 - 27905053 AUGUGGUUACUCGUAUAUUUGCAAUUUCGUUGGCCGGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGUGGCUUUGGCUCCGUGGCCAACUCCGCAUACCCC---- ((((((......(((....)))......((((((((..(((.((((((.......)))))).)))...(..((....))..).)))))))).)))))).....---- ( -35.40) >DroPse_CAF1 100139 98 - 1 GCGUGUGCA-----AUAUUCCCAAUUCUGUUGGCCAGUUUCGAUGGCCUGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGGCUUCAGCUCCAGCUCCAGC---- ((....)).-----..............(((((...(.((((((((((.......)))))))....))).)(((..(((....)))..))).)))))......---- ( -23.70) >DroEre_CAF1 94087 102 - 1 AUGUGGUCACUCGUAUAUUUGCAAUUUCGUUGGCCAGUUUCCAUGGCCCGUU-UUGGCCAUUGAAUGAGUGGCUUUGGCUUCGUGGCCAACUGCAAAGCCCCC---- ....(((((((((((....)))...((((.(((((((...............-))))))).)))).))))))))..(((((.(..(....)..).)))))...---- ( -34.36) >DroYak_CAF1 97613 107 - 1 AUGUGGUCACUCGUAUAUUUGCAAUUUCGUUGGCCAGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGAGGCUUUGGCUUCGUGGCCAACUCCAAAUCCCCCCUGU ...(((......(((....)))......((((((((..(((.((((((.......)))))).)))...(((((....))))).)))))))).)))............ ( -34.90) >DroAna_CAF1 98196 93 - 1 AUGUG-----CCAUAUAUUGGCAAUUUUGUUGGCCAGUUUCCAUGGCCUGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGUGGCCAACUCGGUUU--------- ...((-----(((.....))))).....((((((((......((((((.......))))))..........((....))....)))))))).......--------- ( -31.60) >DroPer_CAF1 97420 98 - 1 GCGUGUGCA-----AUAUUCCCAAUUCUGUUGGCCAGUUUCGAUGGCCUGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGGCUUCAGCUCCAGCUCCAGC---- ((....)).-----..............(((((...(.((((((((((.......)))))))....))).)(((..(((....)))..))).)))))......---- ( -23.70) >consensus AUGUGGUCA_UCGUAUAUUUGCAAUUUCGUUGGCCAGUUUCCAUGGCCCGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGUGGCCAACUCCAAAUCCCCC____ ............................((((((((......((((((.......))))))..........((....))....))))))))................ (-19.68 = -20.15 + 0.47)


| Location | 25,406,141 – 25,406,242 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.23 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -12.70 |
| Energy contribution | -11.98 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25406141 101 + 27905053 GCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACCGGCCAACGAAAUUGCAAAUAUACGAGUAACCACAUACAGUGGGCGCCAGUUUUCU--G-CAUGUUC- (((.........((((((.......))))))(....).)))....(((..((((.....((..((..((((.....))))..))..))....)--)-))..)))- ( -28.00) >DroPse_CAF1 100169 98 + 1 GCGAUUCAUUGAAUGGCCAAAAACAGGCCAUCGAAACUGGCCAACAGAAUUGGGAAUAU-----UGCACACGCACAGGUGACACUGGCUAGCAGAG-AA-AGUCG .(((((..((((.(((((.......)))))))))..(((((((................-----(((....)))..(....)..))))))).....-..-))))) ( -27.00) >DroSim_CAF1 90491 102 + 1 GCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACCGGCCAACGAAAUUGCAAAUAUACGAGUAACCACAUACAGUGGACGCCAGUUUUCCCAG-CA-AUUC- ((..........((((((.......))))))((((((.(((........((((..........))))((((.....))))..))).))))))...)-).-....- ( -25.90) >DroEre_CAF1 94117 97 + 1 GCCACUCAUUCAAUGGCCAA-AACGGGCCAUGGAAACUGGCCAACGAAAUUGCAAAUAUACGAGUGACCACAUACAGUGGGCGCCAGUUUUCCGAGGC------- (((.........((((((..-....))))))(((((((((((..((...(((........))).)).((((.....))))).))))))))))...)))------- ( -33.40) >DroAna_CAF1 98221 97 + 1 GCGAUUCAUUGAAUGGCCAAAAACAGGCCAUGGAAACUGGCCAACAAAAUUGCCAAUAUAUGG-----CACAUACAGUGGUCGCUAGAUGGGAAUG-GA-AUUC- (((((.(((((.(((..........(((((.(....))))))........(((((.....)))-----))))).))))))))))............-..-....- ( -31.90) >DroPer_CAF1 97450 98 + 1 GCGAUUCAUUGAAUGGCCAAAAACAGGCCAUCGAAACUGGCCAACAGAAUUGGGAAUAU-----UGCACACGCACAGGUGACACUGGCUAGCAGAG-AA-AGCCG ((..(((.((((.(((((.......)))))))))..(((((((................-----(((....)))..(....)..)))))))....)-))-.)).. ( -27.70) >consensus GCCACUCAUUCAAUGGCCAAAAACAGGCCAUGGAAACUGGCCAACAAAAUUGCAAAUAUACGA_UGACCACAUACAGUGGACGCCAGUUUGCAGAG_CA_AUUC_ ((((((..((((.(((((.......)))))))))...((.....)).............................))))).)....................... (-12.70 = -11.98 + -0.72)



| Location | 25,406,174 – 25,406,269 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 74.40 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25406174 95 - 27905053 A-AAAAUACUAUAUUUUUUAAUAUAGCUGAAC--AUG-C--AGAAAACUGGCGCCCACUGUAUGUGGUUACUCGUAUAUUUGCAAUUUCGUUGGCCGGUUU .-......(((((((....)))))))(((...--...-)--)).((((((((..((((.....))))......(((....)))..........)))))))) ( -22.30) >DroSec_CAF1 90130 95 - 1 A-AUAAUACUAUAUU-UUAUGUAUAGCUGAAU---UG-CUGGGAAAACUGGCGUCCACUGUAUGUGGUUACUCGUAUAUUUGCAAUUUCGUUGGCCGGUUU .-.............-.......((((.....---.)-)))...((((((((..((((.....))))......(((....)))..........)))))))) ( -20.50) >DroSim_CAF1 90524 96 - 1 A-AUAAUACUAUAUUUUUAUGUAUAGCUGAAU---UG-CUGGGAAAACUGGCGUCCACUGUAUGUGGUUACUCGUAUAUUUGCAAUUUCGUUGGCCGGUUU .-.......(((((....)))))((((.....---.)-)))...((((((((..((((.....))))......(((....)))..........)))))))) ( -20.70) >DroEre_CAF1 94149 92 - 1 A-ACGAUACUUUGGUUUUAUGUGUAGCU--------GCCUCGGAAAACUGGCGCCCACUGUAUGUGGUCACUCGUAUAUUUGCAAUUUCGUUGGCCAGUUU .-......((..(((.(((....)))..--------)))..)).(((((((((.((((.....)))).)....(((....)))..........)))))))) ( -20.80) >DroYak_CAF1 97680 96 - 1 -----AUACUUUAUUUUUAUGUAUAGCUACAUACAUGCAUUGGAAAACUGGCGCCCACUGUAUGUGGUCACUCGUAUAUUUGCAAUUUCGUUGGCCAGUUU -----............(((((((......))))))).......(((((((((.((((.....)))).)....(((....)))..........)))))))) ( -22.30) >DroMoj_CAF1 107520 82 - 1 GUAU-AUAGAGUA-UUUCGUGUA-GA-----U---AU-CCCGCUUAGCUGGCCUUCACUGUAUGUGC-------AGUAUUCACAAAUGCGCUGGCCAGUUU ....-...(((((-(.((.....-))-----.---))-...))))((((((((..(((.....))).-------.(((((....)))))...)))))))). ( -17.70) >consensus A_AUAAUACUAUAUUUUUAUGUAUAGCUGAAU___UG_CUCGGAAAACUGGCGCCCACUGUAUGUGGUUACUCGUAUAUUUGCAAUUUCGUUGGCCAGUUU ............................................((((((((..((((.....))))......(((....)))..........)))))))) (-15.22 = -15.22 + 0.00)


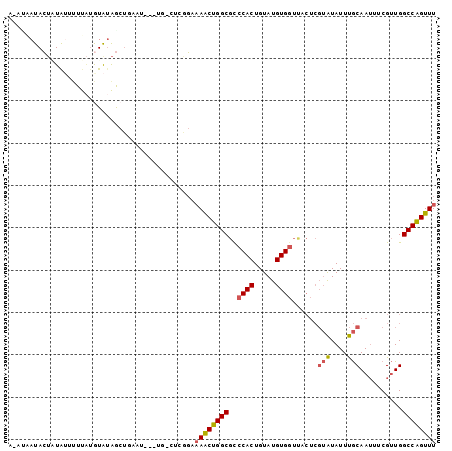
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:27 2006