| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,404,817 – 25,404,913 |
| Length | 96 |
| Max. P | 0.999480 |

| Location | 25,404,817 – 25,404,913 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.41 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -12.99 |
| Energy contribution | -15.00 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.48 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25404817 96 + 27905053 CUAAUGAUUUUGCUCGUUG----UUAUAUG-----AUAUGCCACAUAUAUACAUAUGUACAUAUGUGGUUCCACUAGGAAGAUCUCCAAAUUGACGAGCAAAUUC .....((.(((((((((((----......(-----((..(((((((((.(((....))).)))))))))(((....)))..))).......))))))))))).)) ( -31.12) >DroSim_CAF1 89180 84 + 1 CUAAUGAUUUGGCUCGUUG----UUAUAUG-----AUAUGCCGCAUAU------------AUAGGUGGUUCCACUCGGAACAUCUCCAAAUUGACGAGCAAAUUC .....((((((.((((((.----.((((((-----........)))))------------)..((..(((((....)))))....)).....)))))))))))). ( -23.50) >DroYak_CAF1 96244 92 + 1 CUGCUCAUCU-GCAUGCUGCAUCAAAUACGAAACUCUAGGCUACAGAU------------AGAGGUUGUUCCACUCGGAACAUCUCCAAAUUGAUGAGCAAAUUC .(((((((((-((.....)))........((..(((((..(....).)------------))))..((((((....))))))))........))))))))..... ( -26.10) >consensus CUAAUGAUUU_GCUCGUUG____UUAUAUG_____AUAUGCCACAUAU____________AUAGGUGGUUCCACUCGGAACAUCUCCAAAUUGACGAGCAAAUUC ...........(((((((.....................(((((.(((((((....))))))).))))).......(((.....))).....)))))))...... (-12.99 = -15.00 + 2.01)

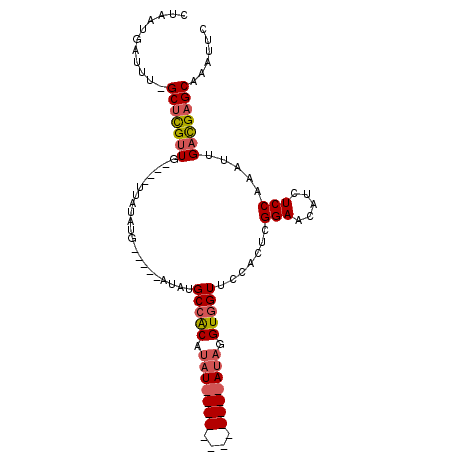

| Location | 25,404,817 – 25,404,913 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 70.41 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -10.65 |
| Energy contribution | -13.77 |
| Covariance contribution | 3.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25404817 96 - 27905053 GAAUUUGCUCGUCAAUUUGGAGAUCUUCCUAGUGGAACCACAUAUGUACAUAUGUAUAUAUGUGGCAUAU-----CAUAUAA----CAACGAGCAAAAUCAUUAG ((.(((((((((.......((....((((....))))((((((((((((....))))))))))))....)-----)......----..))))))))).))..... ( -33.46) >DroSim_CAF1 89180 84 - 1 GAAUUUGCUCGUCAAUUUGGAGAUGUUCCGAGUGGAACCACCUAU------------AUAUGCGGCAUAU-----CAUAUAA----CAACGAGCCAAAUCAUUAG ((.((.((((((.....((..(((((.(((.(((....)))....------------.....))).))))-----).))...----..)))))).)).))..... ( -18.40) >DroYak_CAF1 96244 92 - 1 GAAUUUGCUCAUCAAUUUGGAGAUGUUCCGAGUGGAACAACCUCU------------AUCUGUAGCCUAGAGUUUCGUAUUUGAUGCAGCAUGC-AGAUGAGCAG .....((((((((....(((((.((((((....))))))..))))------------)..(((.((...((((.....))))...)).)))...-.)))))))). ( -28.70) >consensus GAAUUUGCUCGUCAAUUUGGAGAUGUUCCGAGUGGAACCACCUAU____________AUAUGUGGCAUAU_____CAUAUAA____CAACGAGC_AAAUCAUUAG ..(((((((((((.((((((((...)))))))).)).((((.(((((((....))))))).))))........................))))).))))...... (-10.65 = -13.77 + 3.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:23 2006