
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,404,422 – 25,404,553 |
| Length | 131 |
| Max. P | 0.969173 |

| Location | 25,404,422 – 25,404,528 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.75 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -22.91 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25404422 106 + 27905053 AGGCCAAAUACUGGAAUGCCGAGUGUAAGCCAGGAGUCU---CUGAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG .(((...(((((((....)).)))))..)))....(((.---((.....((((......)))).(((((.(((.(((((......)))))))).)))))..)))))... ( -30.70) >DroPse_CAF1 98427 99 + 1 UGUCCAGUCUCUGGGACUCUG----UU-----AGAGCUUC-UCUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUCUGACAAAGGACGGC .((((((((.....)))).((----((-----(((.....-..(((.((((((......)))))))))..(((.(((((......)))))))))))))))..))))... ( -32.30) >DroEre_CAF1 92385 87 + 1 AGGCCAAAUACUG-------------------AGAGCUU---CUGAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG .(.((........-------------------..(((((---.(((....)))..)))))....(((((.(((.(((((......)))))))).)))))...)).)... ( -24.30) >DroWil_CAF1 112191 95 + 1 GGGCCAAAU-----GACUUUG----CU-----UGGGCUUGGGCUGAUCAUUCAUCAAGCUAACUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGGCAAA ..(((....-----.......----..-----..(((((((..(((....))))))))))...((((((.(((.(((((......)))))))).))))))...)))... ( -31.40) >DroYak_CAF1 95785 106 + 1 AGGCCAAAUACUGGAGUGCCGAGUGUAAGCCAAGAGCUU---CUGAUCAUUCAUCAAGCUAAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG .(((...(((((((....)).)))))..)))...(((((---.(((....)))..)))))...((((((.(((.(((((......)))))))).))))))......... ( -32.50) >DroPer_CAF1 95685 99 + 1 UGUCCAGUCUCUGGGACUCUG----UU-----AGAGCUUC-UCUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACGGC .((((((((.....))))...----..-----..(((((.-..(((....)))..)))))...((((((.(((.(((((......)))))))).))))))..))))... ( -31.90) >consensus AGGCCAAAUACUGGGACUCUG____UU_____AGAGCUU___CUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG .(.((.............................(((((....(((....)))..)))))....(((((.(((.(((((......)))))))).)))))...)).)... (-22.91 = -22.97 + 0.06)


| Location | 25,404,422 – 25,404,528 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.75 |
| Mean single sequence MFE | -28.69 |
| Consensus MFE | -19.97 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25404422 106 - 27905053 CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUCAG---AGACUCCUGGCUUACACUCGGCAUUCCAGUAUUUGGCCU .((((((...(((....(((((((......)))))))...((((((((((......)))))).))))---.)))....))...))))..(((.............))). ( -28.92) >DroPse_CAF1 98427 99 - 1 GCCGUCCUUUGUCAGAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAGA-GAAGCUCU-----AA----CAGAGUCCCAGAGACUGGACA ...((((...(((....(((((((......)))))))...((((((((((......)))))).)))).-(..((((.-----..----..))))..)...))).)))). ( -32.00) >DroEre_CAF1 92385 87 - 1 CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUCAG---AAGCUCU-------------------CAGUAUUUGGCCU ..........((((((.(((((((......)))))))...((((((((((......)))))).))))---.......-------------------.....)))))).. ( -22.60) >DroWil_CAF1 112191 95 - 1 UUUGCCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAGUUAGCUUGAUGAAUGAUCAGCCCAAGCCCA-----AG----CAAAGUC-----AUUUGGCCC (((((...(((((((..(((((((......))))).))..)))))))...(((((.(((....)))...)))))...-----.)----))))(((-----....))).. ( -28.10) >DroYak_CAF1 95785 106 - 1 CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUUAGCUUGAUGAAUGAUCAG---AAGCUCUUGGCUUACACUCGGCACUCCAGUAUUUGGCCU .((((((..((((((..(((((((......))))).))..))))))...(((((..(((....))).---)))))...))...))))..(((.(..........)))). ( -28.50) >DroPer_CAF1 95685 99 - 1 GCCGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAGA-GAAGCUCU-----AA----CAGAGUCCCAGAGACUGGACA ...((((...(((....(((((((......)))))))...((((((((((......)))))).)))).-(..((((.-----..----..))))..)...))).)))). ( -32.00) >consensus CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAG___AAGCUCU_____AA____CAGAGUCCCAGUAUUUGGCCU ...(.((..((((((..(((((((......))))).))..))))))...(((((..(((....)))....))))).............................)).). (-19.97 = -19.80 + -0.17)



| Location | 25,404,459 – 25,404,553 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -16.75 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25404459 94 + 27905053 CU--CUGAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGG-ACACGACAAGGCGAUUGCA---GCUAUCGGAAU .(--(((((.........(((((...(((((.(((.(((((......)))))))).))))).....-...((......))....))---))))))))).. ( -26.80) >DroVir_CAF1 105010 84 + 1 UU--AUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGGCAGGCGUUCGCCAUUUGCCGUUUGCC-------------- ..--((((....))))(((((....((((((.(((.(((((......)))))))).))))))(((....))).......)))))..-------------- ( -26.00) >DroPse_CAF1 98455 92 + 1 UUCUCUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUCUGACAAAGG-ACGGCACAAGGCGUUUGCCC---A----AAAAU .....(((.((((((......))))))))).((((..(((((((.....(((((((((.....)))-).))))))))))))...)))---)----..... ( -28.80) >DroYak_CAF1 95822 97 + 1 UU--CUGAUCAUUCAUCAAGCUAAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGG-ACACGACAAGGCGAUUGCAGCAGCUAUCGGUAU ..--(((((.........((((...((((((.(((.(((((......)))))))).))))))....-..........((.......)))))))))))... ( -24.00) >DroAna_CAF1 96392 94 + 1 UU--CUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGG-ACACGACACGGCGUUUGUU---GCUAUCGGAAU ((--(((((....((....((((..((((((.(((.(((((......)))))))).))))))..(.-...)....))))...))..---...))))))). ( -27.80) >DroPer_CAF1 95713 92 + 1 UUCUCUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGG-ACGGCACAAGGCGUUUGCCC---A----AAAAU .....(((.((((((......))))))))).((((..(((((((....(((.(((((....)))))-)))....)))))))...)))---)----..... ( -26.30) >consensus UU__CUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGG_ACACCACAAGGCGUUUGCC____C____GGAAU .....((((.....)))).......((((((.(((.(((((......)))))))).))))))...................................... (-16.75 = -16.95 + 0.20)



| Location | 25,404,459 – 25,404,553 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25404459 94 - 27905053 AUUCCGAUAGC---UGCAAUCGCCUUGUCGUGU-CCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUCAG--AG ....((((((.---.((....)).))))))...-.(((((((((..(((((((......))))).))..))))((((((......))))))..)))--)) ( -24.50) >DroVir_CAF1 105010 84 - 1 --------------GGCAAACGGCAAAUGGCGAACGCCUGCCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAU--AA --------------((.....((((...(((....)))))))....(((((((......)))))))))..(((((((((......)))))).))).--.. ( -28.70) >DroPse_CAF1 98455 92 - 1 AUUUU----U---GGGCAAACGCCUUGUGCCGU-CCUUUGUCAGAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAGAGAA .....----(---((((....(((((.((.((.-....)).)).)))))((((......))))).))))((((((((((......)))))).)))).... ( -28.10) >DroYak_CAF1 95822 97 - 1 AUACCGAUAGCUGCUGCAAUCGCCUUGUCGUGU-CCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUUAGCUUGAUGAAUGAUCAG--AA ....((((.((....)).))))..(..(((.((-....((((((..(((((((......))))).))..))))))....)).)))..)........--.. ( -26.00) >DroAna_CAF1 96392 94 - 1 AUUCCGAUAGC---AACAAACGCCGUGUCGUGU-CCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAG--AA ((((....(((---.....((((......))))-....((((((..(((((((......))))).))..))))))....)))....))))......--.. ( -22.50) >DroPer_CAF1 95713 92 - 1 AUUUU----U---GGGCAAACGCCUUGUGCCGU-CCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAGAGAA .....----(---((((....(((((.((.((.-....)).)).)))))((((......))))).))))((((((((((......)))))).)))).... ( -26.00) >consensus AUUCC____G____GGCAAACGCCUUGUCGCGU_CCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAG__AA ..............................................(((((((......)))))))....(((((((((......)))))).)))..... (-17.41 = -17.13 + -0.28)

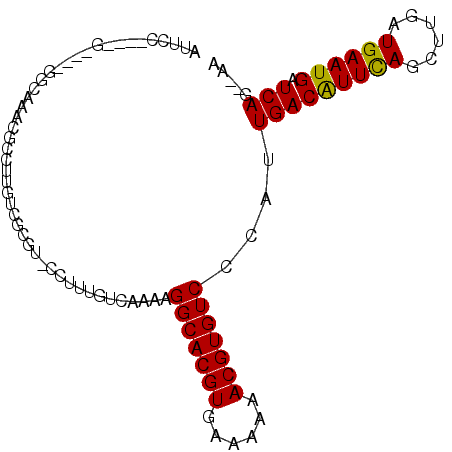

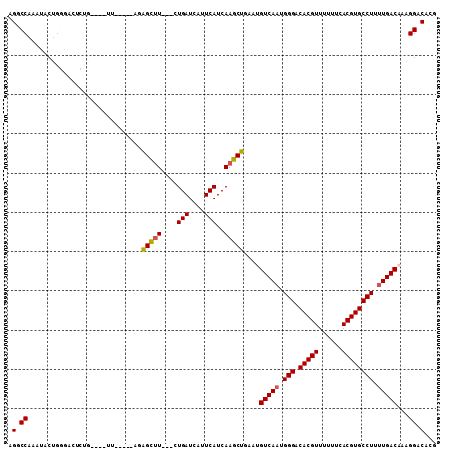
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:22 2006