
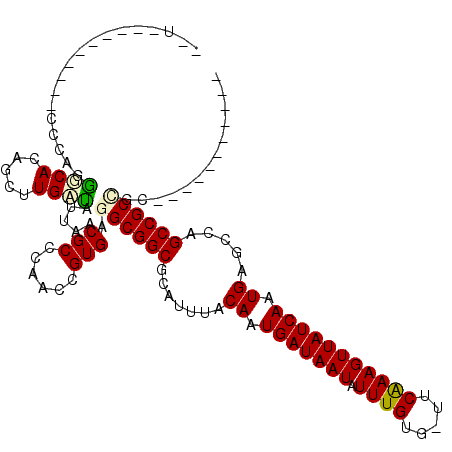
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,393,291 – 25,393,388 |
| Length | 97 |
| Max. P | 0.981265 |

| Location | 25,393,291 – 25,393,388 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25393291 97 + 27905053 -UG----------CCGAAAUCACAGCUUGAUAUUAACGCCCAACUGUGAGGCGGCGCAUUUACAAUGAUAAUAUUUGUG-UUCAAAGUUAUCAAUGGGCCAGCCGCCAA----------- -..----------......((((((.(((...........)))))))))((((((((.....((.(((((((.((((..-..))))))))))).)).))..))))))..----------- ( -30.50) >DroVir_CAF1 92116 102 + 1 A-UA----CUAAGACGGCGGCACAGCAUGCCACUAACGCCCCACCGUGAGGCGGCGCAUUUACAAUGAUAAUAUUUGUG-UCCAAAGUUAUCAAUGAGUGCGCCGC-GU----------- .-..----((...((((.((((.....)))).(....).....)))).))((((((((((((...(((((((.((((..-..))))))))))).))))))))))))-..----------- ( -36.70) >DroPse_CAF1 86350 107 + 1 --U----------CCCAGGUCACAGCUUGAUAUUAACGCCCAACUGUGAGGCGGCGCAUUUACAAUGAUAAUAUUUGUG-UUCAAAGUUAUCAAUGGGCCAGCCGCCGCCGACGACUACG --.----------....((((((((.(((...........)))))))..((((((.......((.(((((((.((((..-..))))))))))).))((....))))))))...))))... ( -32.70) >DroGri_CAF1 87900 104 + 1 ACGA----CGAGAACGGUGGCACAGCAUGAUAUUAACGCCCCACCGUGAGGCGGCGCAUUUACAAUGAUAAUAUUUGUG-UUCGAAGUUAUCAAUGAGUGCGCCGCUGC----------- ....----.....(((((((....((...........)).)))))))..(((((((((((((...(((((((.((((..-..))))))))))).)))))))))))))..----------- ( -39.70) >DroMoj_CAF1 92578 107 + 1 A-UCCCUGCUAGGCGCUGGGCACAGCGUGAUAUUAACGCCCCACCGUGAGGCGGCGCAUUUACAAUGAUAAUAUUUGUGCUCCAAAGUUAUCAAUGAGUGCGCCGC-GC----------- .-......((..(((.((((....((((.......)))))))).))).))((((((((((((...(((((((.((((.....))))))))))).))))))))))))-..----------- ( -41.30) >DroPer_CAF1 83661 107 + 1 --U----------CCCAGGUCACAGCUUGAUAUUAACGCCCAACUGUGAGGCGGCGCAUUUACAAUGAUAAUAUUUGUG-UUCAAAGUUAUCAAUGGGCCAGCCGCCGCCGACGACUACG --.----------....((((((((.(((...........)))))))..((((((.......((.(((((((.((((..-..))))))))))).))((....))))))))...))))... ( -32.70) >consensus __U__________CCCAGGGCACAGCUUGAUAUUAACGCCCAACCGUGAGGCGGCGCAUUUACAAUGAUAAUAUUUGUG_UUCAAAGUUAUCAAUGAGCCAGCCGCCGC___________ ..................((((.....)))).....(((......))).((((((.......((.(((((((.((((.....))))))))))).)).....))))))............. (-20.33 = -20.33 + 0.00)


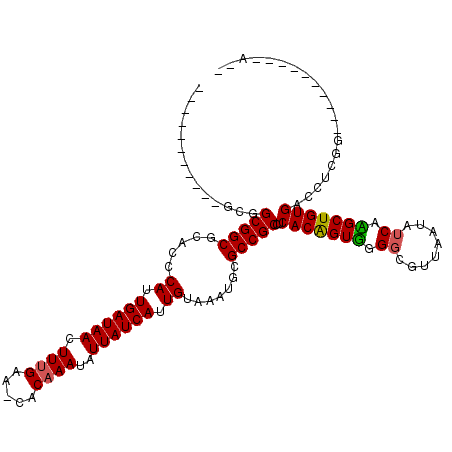
| Location | 25,393,291 – 25,393,388 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -34.11 |
| Consensus MFE | -21.17 |
| Energy contribution | -20.70 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25393291 97 - 27905053 -----------UUGGCGGCUGGCCCAUUGAUAACUUUGAA-CACAAAUAUUAUCAUUGUAAAUGCGCCGCCUCACAGUUGGGCGUUAAUAUCAAGCUGUGAUUUCGG----------CA- -----------..((((((..((.((.((((((.((((..-..))))..)))))).)).....))))))))((((((((.((........)).))))))))......----------..- ( -26.80) >DroVir_CAF1 92116 102 - 1 -----------AC-GCGGCGCACUCAUUGAUAACUUUGGA-CACAAAUAUUAUCAUUGUAAAUGCGCCGCCUCACGGUGGGGCGUUAGUGGCAUGCUGUGCCGCCGUCUUAG----UA-U -----------..-((((((((..((.((((((.((((..-..))))..)))))).))....)))))))).....(.(((((((...(((((((...)))))))))))))).----).-. ( -39.50) >DroPse_CAF1 86350 107 - 1 CGUAGUCGUCGGCGGCGGCUGGCCCAUUGAUAACUUUGAA-CACAAAUAUUAUCAUUGUAAAUGCGCCGCCUCACAGUUGGGCGUUAAUAUCAAGCUGUGACCUGGG----------A-- (.(((.....(((((((.(.....((.((((((.((((..-..))))..)))))).)).....))))))))((((((((.((........)).)))))))).))).)----------.-- ( -32.10) >DroGri_CAF1 87900 104 - 1 -----------GCAGCGGCGCACUCAUUGAUAACUUCGAA-CACAAAUAUUAUCAUUGUAAAUGCGCCGCCUCACGGUGGGGCGUUAAUAUCAUGCUGUGCCACCGUUCUCG----UCGU -----------...((((((((..((.((((((.......-........)))))).))....))))))))...(((((((.(((.((......)).))).))))))).....----.... ( -34.76) >DroMoj_CAF1 92578 107 - 1 -----------GC-GCGGCGCACUCAUUGAUAACUUUGGAGCACAAAUAUUAUCAUUGUAAAUGCGCCGCCUCACGGUGGGGCGUUAAUAUCACGCUGUGCCCAGCGCCUAGCAGGGA-U -----------..-((((((((..((.((((((.((((.....))))..)))))).))....))))))))(((..((((.(((((.......))))).))))..((.....)).))).-. ( -39.40) >DroPer_CAF1 83661 107 - 1 CGUAGUCGUCGGCGGCGGCUGGCCCAUUGAUAACUUUGAA-CACAAAUAUUAUCAUUGUAAAUGCGCCGCCUCACAGUUGGGCGUUAAUAUCAAGCUGUGACCUGGG----------A-- (.(((.....(((((((.(.....((.((((((.((((..-..))))..)))))).)).....))))))))((((((((.((........)).)))))))).))).)----------.-- ( -32.10) >consensus ___________GCGGCGGCGCACCCAUUGAUAACUUUGAA_CACAAAUAUUAUCAUUGUAAAUGCGCCGCCUCACAGUGGGGCGUUAAUAUCAAGCUGUGACCUCGG__________A__ ..............(((((.....((.((((((.((((.....))))..)))))).)).......)))))..(((((((.((........)).))))))).................... (-21.17 = -20.70 + -0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:14 2006