
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,350,613 – 25,350,732 |
| Length | 119 |
| Max. P | 0.968303 |

| Location | 25,350,613 – 25,350,732 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -18.60 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25350613 119 + 27905053 AAGUCAUUCCUCUUAAUAGUCAUGCAAAUUAAUUUUUGCUCAUUUGUACAACAAUAAAAAUGUUAACUAAUUAAUUUUGAUUGAAAUUUGCAUGCAAAACGCGAAAAUCAAUUUGCACG ......................((((((((.((((((((...(((((((((((((.(((((............))))).))))....)))..))))))..)))))))).)))))))).. ( -20.80) >DroVir_CAF1 39446 118 + 1 AAGUCAUUCCUUUUAAUAGUCAUGCAAAUUAAUU-UUGCUCAUUUGUACAACAAUAAAAAUGUUAACUAAUUAAUUUUGAUUGAAAUUUGCAUGCAAAGAGCGAAAAUCAAUUUGCACG ......................((((((((..((-((((((.(((((((((((((.(((((............))))).))))....)))..))))))))))))))...)))))))).. ( -22.60) >DroGri_CAF1 36239 118 + 1 AAGUCAUUCCUUUUAAUAGUCAUGCAAAUUAAUU-UUGCUCAUUUGUACAACAAUAAAAAUGUUAACUAAUUAAUUUUGAUUGAAAUUUGCAUGCAAAGAGCGAAAAUCAAUUUGCAUG ....................((((((((((..((-((((((.(((((((((((((.(((((............))))).))))....)))..))))))))))))))...)))))))))) ( -25.60) >DroEre_CAF1 36170 119 + 1 AAGUCAUUCCUCUUAAUAGUCAUGCAAAUUAAUUUUUGCUCAUUUGUACAACAAUAAAAAUGUUAACUAAUUAAUUUUGAUUGAAAUUUGCAUGCGAAACGCGAAAAUCAAUUUGCACG ......................((((((((.((((((((...(((((((((((((.(((((............))))).))))....)))..))))))..)))))))).)))))))).. ( -20.50) >DroMoj_CAF1 38834 118 + 1 AAGUCAUUCCUUUUAAUAGUCAUGCAAAUUAAUU-UUGCUCAUUUGUACAACAAUAAAAAUGUUAACUAAUUAAUUUUGAUUGAAAUUUGCAUGCAAAGAGCGAAAAUCAAUUUGCACG ......................((((((((..((-((((((.(((((((((((((.(((((............))))).))))....)))..))))))))))))))...)))))))).. ( -22.60) >DroAna_CAF1 41674 119 + 1 AAGUCAUUCCUUUUAAUAGUCAUGCAAAUUAAUUUUUGCUCAUUUGUACAACAAUAAAAAUGUUAACUAAUUAAUUUUGAUUGAAAUUUGCAUGCAAAAUGCGAAAAUCAAUUUGCACG ......................((((((((.((((((((...(((((((((((((.(((((............))))).))))....)))..))))))..)))))))).)))))))).. ( -21.50) >consensus AAGUCAUUCCUUUUAAUAGUCAUGCAAAUUAAUU_UUGCUCAUUUGUACAACAAUAAAAAUGUUAACUAAUUAAUUUUGAUUGAAAUUUGCAUGCAAAAAGCGAAAAUCAAUUUGCACG ..................(.((((((((((......(((......)))...((((.(((((............))))).)))).))))))))))).....(((((......)))))... (-18.60 = -18.60 + -0.00)



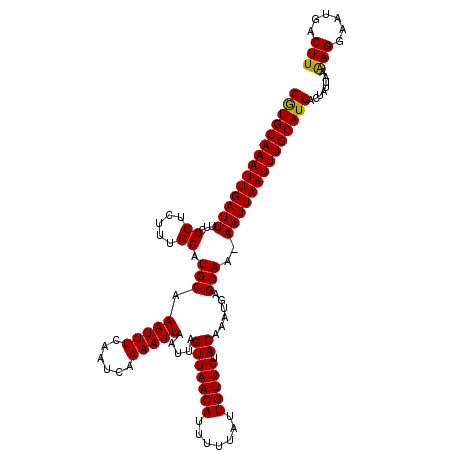
| Location | 25,350,613 – 25,350,732 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -22.23 |
| Energy contribution | -21.87 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25350613 119 - 27905053 CGUGCAAAUUGAUUUUCGCGUUUUGCAUGCAAAUUUCAAUCAAAAUUAAUUAGUUAACAUUUUUAUUGUUGUACAAAUGAGCAAAAAUUAAUUUGCAUGACUAUUAAGAGGAAUGACUU ((((((((((((((((.((..((((..(((((....((((.(((((............))))).)))))))))))))...)).))))))))))))))))........(((......))) ( -25.30) >DroVir_CAF1 39446 118 - 1 CGUGCAAAUUGAUUUUCGCUCUUUGCAUGCAAAUUUCAAUCAAAAUUAAUUAGUUAACAUUUUUAUUGUUGUACAAAUGAGCAA-AAUUAAUUUGCAUGACUAUUAAAAGGAAUGACUU ((((((((((((((((.((((((((..(((((....((((.(((((............))))).))))))))))))).))))))-)))))))))))))).................... ( -28.50) >DroGri_CAF1 36239 118 - 1 CAUGCAAAUUGAUUUUCGCUCUUUGCAUGCAAAUUUCAAUCAAAAUUAAUUAGUUAACAUUUUUAUUGUUGUACAAAUGAGCAA-AAUUAAUUUGCAUGACUAUUAAAAGGAAUGACUU ((((((((((((((((.((((((((..(((((....((((.(((((............))))).))))))))))))).))))))-)))))))))))))).................... ( -28.90) >DroEre_CAF1 36170 119 - 1 CGUGCAAAUUGAUUUUCGCGUUUCGCAUGCAAAUUUCAAUCAAAAUUAAUUAGUUAACAUUUUUAUUGUUGUACAAAUGAGCAAAAAUUAAUUUGCAUGACUAUUAAGAGGAAUGACUU ((((((((((((((((.(((...))).(((.(((((......))))).....(((((((.......))))).))......)))))))))))))))))))........(((......))) ( -24.50) >DroMoj_CAF1 38834 118 - 1 CGUGCAAAUUGAUUUUCGCUCUUUGCAUGCAAAUUUCAAUCAAAAUUAAUUAGUUAACAUUUUUAUUGUUGUACAAAUGAGCAA-AAUUAAUUUGCAUGACUAUUAAAAGGAAUGACUU ((((((((((((((((.((((((((..(((((....((((.(((((............))))).))))))))))))).))))))-)))))))))))))).................... ( -28.50) >DroAna_CAF1 41674 119 - 1 CGUGCAAAUUGAUUUUCGCAUUUUGCAUGCAAAUUUCAAUCAAAAUUAAUUAGUUAACAUUUUUAUUGUUGUACAAAUGAGCAAAAAUUAAUUUGCAUGACUAUUAAAAGGAAUGACUU ((((((((((((((((.((..((((....))))...................(((((((.......))))).))......)).)))))))))))))))).................... ( -25.00) >consensus CGUGCAAAUUGAUUUUCGCUCUUUGCAUGCAAAUUUCAAUCAAAAUUAAUUAGUUAACAUUUUUAUUGUUGUACAAAUGAGCAA_AAUUAAUUUGCAUGACUAUUAAAAGGAAUGACUU ((((((((((((((...((.....)).(((.(((((......))))).....(((((((.......))))).))......)))..))))))))))))))........(((......))) (-22.23 = -21.87 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:37:57 2006