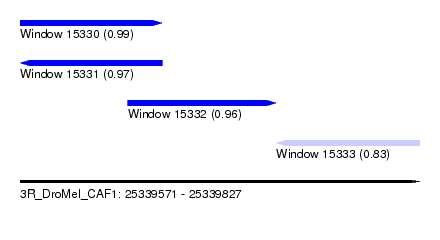
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,339,571 – 25,339,827 |
| Length | 256 |
| Max. P | 0.994146 |

| Location | 25,339,571 – 25,339,662 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25339571 91 + 27905053 CGGCUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCCAGCGACGAUGUCACCGG-----------------GCAGCAACAAUUG---------CAGUUGC---UG ..(((((((...(((((((((((...((((((....)))))).........))))))))))).....))-----------------))))).......(---------(....))---.. ( -36.60) >DroPse_CAF1 26276 87 + 1 CGCUUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCAAGCGACGAUGUCAGC------------------------CACAG------CCACUGCCACUGC---CA .(((((.((...((((((((((....((((((....))))))..........))))))))))..)).------------------------)).))------)............---.. ( -27.04) >DroSec_CAF1 24539 100 + 1 CGGCUGUCUCAUCUUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCCAGCGACGAUGUCACCGG-----------------GCAGCAACAAUUGCAGCAGUUGCAGUUGC---UC ..(((((((...(.(((((((((...((((((....)))))).........))))))))).).....))-----------------)))))..(((((((((...))))))))).---.. ( -39.80) >DroSim_CAF1 24924 100 + 1 CGGCUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCCAGCGACGAUGUCACCGG-----------------GCAGCAACAAUUGCAGCAGUUGCAGUUGC---UG ..(((((((...(((((((((((...((((((....)))))).........))))))))))).....))-----------------)))))..(((((((((...))))))))).---.. ( -43.70) >DroAna_CAF1 30450 120 + 1 CGCCUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCAAGCGACGAUGUCACCAACUGCAAAUGCAACUGCAACAUCACCAGCCGAAGCAGAAGCAGAAGCAGAAG ............((((((((((....((((((....))))))..........)))))))))).......((((...(((..((((..(..........)..))))..)))...))))... ( -37.24) >DroPer_CAF1 25785 93 + 1 CGCUUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCAAGCGACGAUGUCAGC------------------------CACAGCCACAGCCACUGCCACUGC---CA .(((((.((...((((((((((....((((((....))))))..........))))))))))..)).------------------------)).)))..................---.. ( -27.04) >consensus CGCCUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCAAGCGACGAUGUCACCGG_________________GCAGCAACAAUUGCAGCAGCUGCAGUUGC___UG .((.((......((((((((((....((((((....))))))..........)))))))))).))))..................................................... (-21.84 = -21.84 + -0.00)


| Location | 25,339,571 – 25,339,662 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -21.99 |
| Energy contribution | -21.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25339571 91 - 27905053 CA---GCAACUG---------CAAUUGUUGCUGC-----------------CCGGUGACAUCGUCGCUGGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAGCCG ..---((....)---------)..((((((((((-----------------((((((((...))))))))).......((((((....))))))...))))))))).............. ( -36.60) >DroPse_CAF1 26276 87 - 1 UG---GCAGUGGCAGUGG------CUGUG------------------------GCUGACAUCGUCGCUUGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAAGCG ((---.((((.((((...------)))).------------------------)))).))....((((((........((((((....))))))((((.........))))...)))))) ( -29.40) >DroSec_CAF1 24539 100 - 1 GA---GCAACUGCAACUGCUGCAAUUGUUGCUGC-----------------CCGGUGACAUCGUCGCUGGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAAAGAUGAGACAGCCG (.---((...((((.....)))).((((((((((-----------------((((((((...))))))))).......((((((....))))))...)))))))))..........))). ( -40.20) >DroSim_CAF1 24924 100 - 1 CA---GCAACUGCAACUGCUGCAAUUGUUGCUGC-----------------CCGGUGACAUCGUCGCUGGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAGCCG ((---(((........)))))...((((((((((-----------------((((((((...))))))))).......((((((....))))))...))))))))).............. ( -40.10) >DroAna_CAF1 30450 120 - 1 CUUCUGCUUCUGCUUCUGCUUCGGCUGGUGAUGUUGCAGUUGCAUUUGCAGUUGGUGACAUCGUCGCUUGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAGGCG ..((((..((.(((...((....)).)))(((((..(..((((....))))...)..)))))((((((.((.......((((((....))))))..))))))))......))..)))).. ( -41.40) >DroPer_CAF1 25785 93 - 1 UG---GCAGUGGCAGUGGCUGUGGCUGUG------------------------GCUGACAUCGUCGCUUGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAAGCG ..---(((((.((((...)))).))))).------------------------(((....((((((.(((.(((....((((((....))))))....)))..)))))))))....))). ( -35.70) >consensus CA___GCAACUGCAACUGCUGCAACUGUUGCUGC_________________CCGGUGACAUCGUCGCUGGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAGCCG .....((....))........................................(((..((((((((((.((.......((((((....))))))..))))))))...)))).....))). (-21.99 = -21.77 + -0.22)


| Location | 25,339,640 – 25,339,735 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -20.46 |
| Energy contribution | -21.78 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25339640 95 + 27905053 GCAGCAACAAUUG---------CAGUUGCUGUUGCAGUUGCAGCAGCGAUGACGACGAAUUAUAGCAAAGUAAUCGCAGUGGACUCUCCAGUUGCCAGUGCAUC ((((((((.((((---------(.(((((((((((....)))))))))))......((.((((......)))))))))))((.....)).)))))...)))... ( -33.70) >DroPse_CAF1 26343 83 + 1 -----CACAG------CCACUGCCACUGCCACAGCAGCAGCUCCAGCGAUGACGACGAAUUAUAGCAAAGUAAUCGCACUGGACUCUGUAGU----------GC -----.....------.((((((..((((....)))).((.(((((((((.((................)).)))))..))))))..)))))----------). ( -20.59) >DroSec_CAF1 24608 104 + 1 GCAGCAACAAUUGCAGCAGUUGCAGUUGCUCUUGCAGUUGCAGCAGCGAUGACGACGAAUUAUAGCAAAGUAAUCGCAGUGGACUCUCCAGUUGCCAGUGCAUC ((((((((.(((((.((.((((((..(((....)))..)))))).))(((.((................)).))))))))((.....)).)))))...)))... ( -34.39) >DroSim_CAF1 24993 104 + 1 GCAGCAACAAUUGCAGCAGUUGCAGUUGCUGUUGCAGUUGCAGCAGCGAUGACGACGAAUUAUAGCAAAGUAAUCGCAGUGGACUCUCCAGUUGCCAGUGCAUC ((((((((.(((((....((((..(((((((((((....)))))))))))..))))((.((((......)))))))))))((.....)).)))))...)))... ( -39.20) >consensus GCAGCAACAAUUG___CAGUUGCAGUUGCUGUUGCAGUUGCAGCAGCGAUGACGACGAAUUAUAGCAAAGUAAUCGCAGUGGACUCUCCAGUUGCCAGUGCAUC ((((((((................(((((((((((....)))))))))))...(.(((.((((......))))))))..(((.....))))))))...)))... (-20.46 = -21.78 + 1.31)



| Location | 25,339,735 – 25,339,827 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 71.34 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -11.14 |
| Energy contribution | -12.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25339735 92 - 27905053 ---------GAUAU-GAUUGUAAUACUUAUAUAUA-AAUGGUAGUUCAGAGUUGGCUUUGGCAUAUCCUACUCCUUGAAAGGGUAUGAUCCAGGUGGACAAAG ---------.....-..(((...((((........-...))))...))).(((.((((.((((((((((..........)))))))).)).)))).))).... ( -19.10) >DroSec_CAF1 24712 95 - 1 AUUG-----CAUAU-AUUUAUAAUACUUAUAUAGC-AA-GGUAAAUCAAAAUUGGCUUUGGCAUAUCCUCUUCCUUGAAAGGGUAUUAUCAAGGUGGACAAAG .(((-----(((((-(...........))))).))-))-............((.(((((((.(((((((.(((...))))))))))..))))))).))..... ( -17.70) >DroSim_CAF1 25097 95 - 1 AUUG-----CAUAU-AUUUAUAAUACUUAUAUAUC-AA-GGUAUAUAAGAGUUGGCUUUGGCAUAUCCUACUCCUUGAAAGGGUAUUAUCAAGGUGGACAAAG ....-----.....-..........(((((((((.-..-.))))))))).(((.(((((((.(((((((..........)))))))..))))))).))).... ( -24.40) >DroEre_CAF1 24943 95 - 1 GCUGCAUUUAAUAA-AUU--UUAUACUG---AAUAAAU-GGUAAACUAGCGCUAC-UUCGGUAUAUCCUGCUCCUCGAAAGGGUAUGCUCGAGGUGGACAAAG ((((..((((....-..(--((((....---.))))).-..)))).))))(((((-(((((((((((((..........))))))))).)))))))).).... ( -26.20) >DroYak_CAF1 27380 89 - 1 AUUG-----AAUGGCAUU--U--UACUU---UAUU-AG-GGUAAAGUAGAGUUAAUUUUGGUAUAUCGUCUUCCUGGAAAGGGUAUGCCUGAGCUGGACAAAG .(((-----..(((((((--(--(((((---(((.-..-.)))))))))))).......((((((((.(((....)))...))))))))...))))..))).. ( -24.80) >consensus AUUG_____AAUAU_AUUU_UAAUACUUAUAUAUA_AA_GGUAAAUCAGAGUUGGCUUUGGCAUAUCCUACUCCUUGAAAGGGUAUGAUCAAGGUGGACAAAG ..................................................(((.(((((((((((((((..........)))))))).))))))).))).... (-11.14 = -12.14 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:37:48 2006