
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,327,258 – 25,327,393 |
| Length | 135 |
| Max. P | 0.738802 |

| Location | 25,327,258 – 25,327,353 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
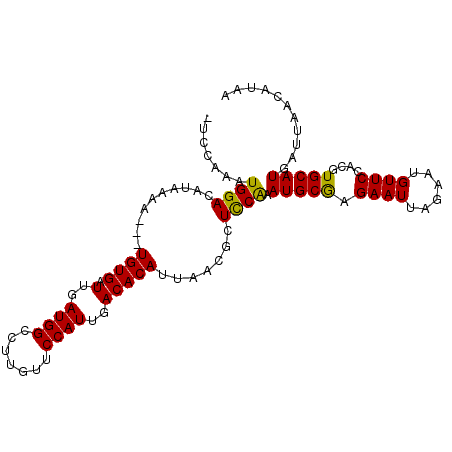
>3R_DroMel_CAF1 25327258 95 + 27905053 AUCCGUUUGCUUCGGCCAUUAAUAACAUAAGUCUCGGAAUGGAUAUUCAAUUCUGUGAUUGAUGGCCUUGUUCCAUUGACACAUUAACGCUCCAA ........((...((((((((.........(((.((((((.(.....).)))))).))))))))))).((((.....)))).......))..... ( -20.30) >DroSec_CAF1 12238 91 + 1 AUCUGUUUGCUUCGGCCAUUAAUAACAGAAGA-UCCAAAUGGACAUAAAA---UGUGAUUGAUGGCCUUGUUCCAUUGACACAUUAACGCUCCAA .((((((.((....)).......))))))...-......((((.....((---((((.(..((((.......))))..))))))).....)))). ( -22.90) >DroSim_CAF1 12209 91 + 1 AUCUGUUUGCUUCGGCCAUUAAUAACAGAAGA-UCCAAAUGGACAUAAAA---UGUGAUUGAUGGCCUUGUUCCAUUGACACAUUAACGCUUCAA ...((...((...((((((((((.(((.....-(((....))).......---))).)))))))))).((((.....)))).......))..)). ( -21.60) >DroYak_CAF1 13112 91 + 1 AUCUGUGCGCUGCGGCCACUAAUAGGAGAGUA-UCUACAUGGACACACAA---UGUGAUUGAUGGCCUUGUGCCAUUGACACAUUAACGCUCCGA ....(((.((....))))).....((((.((.-(((....))).))..((---((((.(..(((((.....)))))..)))))))....)))).. ( -28.10) >consensus AUCUGUUUGCUUCGGCCAUUAAUAACAGAAGA_UCCAAAUGGACAUAAAA___UGUGAUUGAUGGCCUUGUUCCAUUGACACAUUAACGCUCCAA ........((...((((((((((..........(((....)))((((......)))))))))))))).((((.....)))).......))..... (-15.80 = -16.30 + 0.50)



| Location | 25,327,290 – 25,327,393 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.15 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25327290 103 + 27905053 CUCGGAAUGGAUAUUCAAUUCUGUGAUUGAUGGCCUUGUUCCAUUGACACAUUAACGCUCCAAAUGCAAGAAUUAGAAUGUUCCACGUGCAUGAUUAACAUAA .......((((((((((((((((((.(..((((.......))))..))))......((.......)).)))))).))))).)))).................. ( -22.60) >DroSec_CAF1 12270 99 + 1 -UCCAAAUGGACAUAAAA---UGUGAUUGAUGGCCUUGUUCCAUUGACACAUUAACGCUCCAAAUGCGAGAAUUAGAAUGUUCCACGUGCAUGAUUAACAUAA -..((..(((((((..((---((((.(..((((.......))))..)))))))..(((.......))).........))).))))..)).............. ( -21.60) >DroSim_CAF1 12241 99 + 1 -UCCAAAUGGACAUAAAA---UGUGAUUGAUGGCCUUGUUCCAUUGACACAUUAACGCUUCAAAUGCGAGAAUUAGAAUGUUCCACGUGCAUGAUUAACAUAA -..((..(((((((..((---((((.(..((((.......))))..)))))))..(((.......))).........))).))))..)).............. ( -21.60) >DroYak_CAF1 13144 99 + 1 -UCUACAUGGACACACAA---UGUGAUUGAUGGCCUUGUGCCAUUGACACAUUAACGCUCCGAAUGCGAGAAUUAGAAGGUUCCACGUGCAUGAUUAACAUAA -....((((.((....((---((((.(..(((((.....)))))..)))))))..(((.......))).(((((....)))))...)).)))).......... ( -27.00) >consensus _UCCAAAUGGACAUAAAA___UGUGAUUGAUGGCCUUGUUCCAUUGACACAUUAACGCUCCAAAUGCGAGAAUUAGAAUGUUCCACGUGCAUGAUUAACAUAA .......((((..........((((.(..((((.......))))..))))).......)))).(((((.((((......))))....)))))........... (-17.72 = -17.15 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:37:40 2006