
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,288,131 – 25,288,243 |
| Length | 112 |
| Max. P | 0.974568 |

| Location | 25,288,131 – 25,288,243 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.71 |
| Mean single sequence MFE | -51.50 |
| Consensus MFE | -25.31 |
| Energy contribution | -26.06 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25288131 112 - 27905053 UUCCGUUUCCCAUUUCGCCGGCCACCGGAACC-----GG-AAACGCAUUGGCGAAGGCCAACGUGGUGGCUGGCACAUCCGGCGGCUGAUCCUGCUGCUCAAUGGAGGUGGCUACGGU ..((((...(((((((((((((((((......-----(.-...)((.(((((....))))).)))))))))))).(((..((((((.......))))))..))))))))))..)))). ( -60.40) >DroPse_CAF1 151950 118 - 1 AACCAUUGCCCAUUUCCCCGGCCACGGGAACGGGCCCGGGCACGGCGUUGGCGAAGGACAGCGUGGUGGCCGGCACCUCUGGCGGCUGAUCCUGCUGCUCGAUGGAGGUGGGAGAUGU ..........((((((((.((((.((....))))))(((.(((.((((((.(....).)))))).))).)))..((((((((((((.......))))))....)))))))))))))). ( -61.50) >DroGri_CAF1 281047 102 - 1 GUCCAUU----GUAUCGCCGGCAACGGCAA------------CGGCAUUGGCAAAGGCCAAUGUUGUGGCGGGAAUUGCAGCUGGCUGAUCCUGUUGCUCGAUGGAUGUGGGUGGUGU (((((((----(....((((....))))..------------.(((((((((....)))))))))(..((((((....(((....))).))))))..).))))))))........... ( -46.40) >DroEre_CAF1 162519 107 - 1 -----UUGCCCAUUUCGCCGGCCACCGGAACC-----GG-AAACGCAUUUGCGAAGGCCAACGUGGUGGCGGGCACGUCCGGUGGCUGAUCCUGCUGCUCAAUGGAGGUGGCUGCGGU -----..((((((..((..((((.(((....)-----))-...(((....)))..))))..))..)))((((.(((.(((((..((.......))..))....))).))).))))))) ( -47.00) >DroMoj_CAF1 188288 102 - 1 GUCCAUG----GUAUCGCCCGCAACGGCAA------------CUGCAUUGGCAAAGGCCAGGGUGGUGGCAGGGAUCGCCGGCGGCUGGUCCUGCUGCUCAAUGGAAGUGGGUGCCGU ((((.((----.(((((((((((..(....------------))))..((((....)))))))))))).)).)))).....(((((....((..((.((....)).))..)).))))) ( -43.80) >DroAna_CAF1 140542 112 - 1 UGCCAUUGCCCAUUUCCCCCGCCACCGGAACC-----GG-AACUGCGUUGGCAAAGGCCAGUGUGGUGGCCGGCACAUCCGGUGGCUGAUCCUGCUGCUCGAUCGAGGUGGCUGAAGU .(((((..(...........(((((((((.((-----((-.((..(((((((....)))))))..))..))))....))))))))).((((.........)))))..)))))...... ( -49.90) >consensus GUCCAUUGCCCAUUUCGCCGGCCACCGGAACC_____GG_AACCGCAUUGGCAAAGGCCAACGUGGUGGCCGGCACAUCCGGCGGCUGAUCCUGCUGCUCAAUGGAGGUGGCUGCGGU .........(((((((.((.((((((..................((.(((((....))))).)))))))).)).......((((((.......)))))).....)))))))....... (-25.31 = -26.06 + 0.75)

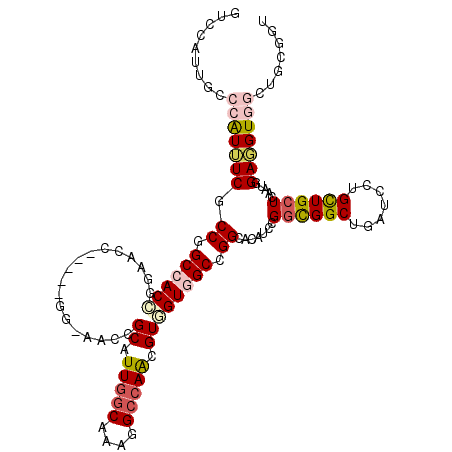

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:37:26 2006