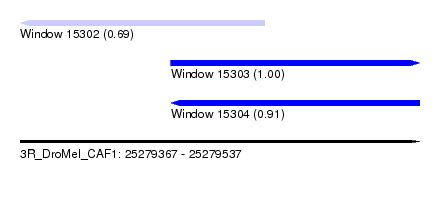
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,279,367 – 25,279,537 |
| Length | 170 |
| Max. P | 0.995460 |

| Location | 25,279,367 – 25,279,471 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.69 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.97 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25279367 104 - 27905053 AGUAGAGCUUCUCCCAAAGCAAACCCGAUUACUCGGUAUUAUCCUGAUUUGAGCUAAUGCUCGUUUGUGAACCCAACCUCCGCAUGGCCUUCGGGCCAACAAAU .(..(((...........((((((((((....))))..............((((....)))))))))).........)))..).(((((....)))))...... ( -31.15) >DroSec_CAF1 136246 99 - 1 AGUAGAGCUGCUCCCA-----AACACGAUUACUCGGCAUUAUCCUGAUUUGAGCUAAUGCUCGUUUGCGAAUCCAGCCUCCGCAUGGCCUUCGGGCCAACAAAU .(..((((((.((.((-----(((.(((....)))...............((((....))))))))).))...))).)))..).(((((....)))))...... ( -33.30) >DroSim_CAF1 135976 104 - 1 AGUAGAGCUGCUCCCAAAGCAAACCCGAUUACUCGGCAUUAUCCUGAUUUGAGUUAAUGCUCGUUUGCGAAUCCAACCUCCGCAUGGCCUUCGGGCCAACAAAU .(..(((...........((((((((((....))))..............((((....)))))))))).........)))..).(((((....)))))...... ( -30.45) >DroEre_CAF1 153632 86 - 1 AGUAAAGCAGCUCCAAUCGCAAACCCGAUUACUCGGCAUUAUCCUGAUUUGAGCUAAUGCU------------------CCGGAUGGCCUUCCGGGCAACAAAU ........(((((.(((((.....((((....))))........))))).)))))..(((.------------------(((((......))))))))...... ( -25.72) >DroYak_CAF1 141592 104 - 1 AGUGAAGCUUCUCCAUGAGCAAACCCGAUUACUUGGCAUUAUCCUGAUUUGAGCUAAUGCUCGUUUGGAAACCCAACCUCCGAAUGGCCUUCCGGGCAACAAAU ......((((......))))...((((.......(((.............((((....))))(((((((.........))))))).)))...))))........ ( -24.50) >consensus AGUAGAGCUGCUCCCAAAGCAAACCCGAUUACUCGGCAUUAUCCUGAUUUGAGCUAAUGCUCGUUUGCGAACCCAACCUCCGCAUGGCCUUCGGGCCAACAAAU ....((((.((((...........((((....))))..............))))....))))......................(((((....)))))...... (-14.81 = -15.97 + 1.16)


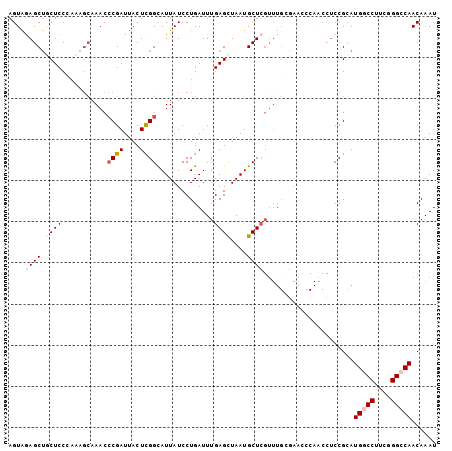
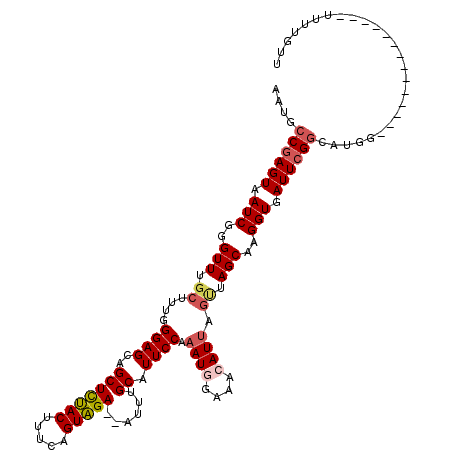
| Location | 25,279,431 – 25,279,537 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25279431 106 + 27905053 AAUACCGAGUAAUCGGGUUUGCUUUGGGAGAAGCUCUACUUUCAGUAGAGCAUUUGCAUUCCAAAUGGAAACAUUAGCUAGCCAGGUGAUUCGGCAUGG--------------UUUUGUU ....((((((.(((.((((.(((.(((((((((((((((.....))))))).)))...)))))((((....))))))).)))).))).)))))).....--------------....... ( -36.80) >DroSec_CAF1 136310 99 + 1 AAUGCCGAGUAAUCGUGUU-----UGGGAGCAGCUCUACUUUCAGUAGA--CUUUGCAUUCCAAAUGGAAACAUUAGUUAGCAAGGUGAUUCGGCAUGG--------------UUUUCUU .(((((((((.(((.((((-----(((((((((.(((((.....)))))--..)))).)))))((((....))))....)))).))).)))))))))..--------------....... ( -35.60) >DroSim_CAF1 136040 104 + 1 AAUGCCGAGUAAUCGGGUUUGCUUUGGGAGCAGCUCUACUUUCAGUAGA--CUUUGCAUUCCAAAUGGAAACAUUAGUUAGCAAGGUGAUUCGGCAUGG--------------UUUUGUU .(((((((((.(((..(((.(((.(((((((((.(((((.....)))))--..)))).)))))((((....))))))).)))..))).)))))))))..--------------....... ( -38.20) >DroEre_CAF1 153678 113 + 1 AAUGCCGAGUAAUCGGGUUUGCGAUUGGAGCUGCUUUACUACCAGUAGA--GUUUGCAUUCCGAAUUGAAACAUCAGUUAGCAAGGUGAUUCGGGAUGAUUCCCUCGCAAUGUU-C---- ....((((....))))..(((((((((((((.(((((((.....)))))--))..))..)))))...(((.((((.........)))).)))((((....))))))))))....-.---- ( -36.50) >DroYak_CAF1 141656 118 + 1 AAUGCCAAGUAAUCGGGUUUGCUCAUGGAGAAGCUUCACUUUCUGUGGA--AUUUGCAUUCCAAAUUGAAACAUUAGUUAGCAAGGUGAUUCGAGAGGGAUGCCUUACUAUGAUUUUGUU ........((((......))))((((((.(((..(((((.....)))))--.)))(((((((...(((((.((((.........)))).)))))..)))))))....))))))....... ( -26.10) >consensus AAUGCCGAGUAAUCGGGUUUGCUUUGGGAGCAGCUCUACUUUCAGUAGA__AUUUGCAUUCCAAAUGGAAACAUUAGUUAGCAAGGUGAUUCGGCAUGG______________UUUUGUU ....((((((.(((..(((.((....((((..(((((((.....)))))......)).)))).((((....)))).)).)))..))).)))))).......................... (-17.00 = -17.72 + 0.72)


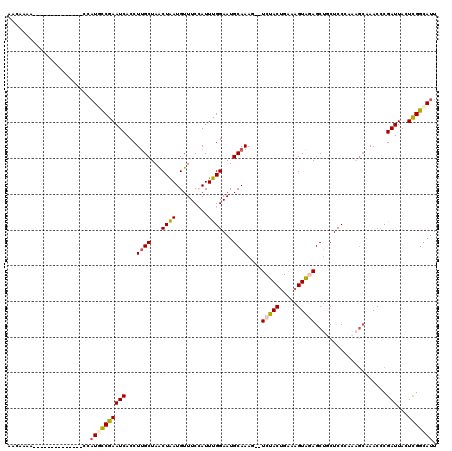
| Location | 25,279,431 – 25,279,537 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -14.40 |
| Energy contribution | -14.56 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25279431 106 - 27905053 AACAAAA--------------CCAUGCCGAAUCACCUGGCUAGCUAAUGUUUCCAUUUGGAAUGCAAAUGCUCUACUGAAAGUAGAGCUUCUCCCAAAGCAAACCCGAUUACUCGGUAUU .......--------------..(((((((.......((...(((..(((((((....)))).)))...(((((((.....))))))).........)))...)).......))))))). ( -27.34) >DroSec_CAF1 136310 99 - 1 AAGAAAA--------------CCAUGCCGAAUCACCUUGCUAACUAAUGUUUCCAUUUGGAAUGCAAAG--UCUACUGAAAGUAGAGCUGCUCCCA-----AACACGAUUACUCGGCAUU .......--------------..((((((((((....((..(((....)))..))(((((...(((...--(((((.....)))))..)))..)))-----))...)))...))))))). ( -24.00) >DroSim_CAF1 136040 104 - 1 AACAAAA--------------CCAUGCCGAAUCACCUUGCUAACUAAUGUUUCCAUUUGGAAUGCAAAG--UCUACUGAAAGUAGAGCUGCUCCCAAAGCAAACCCGAUUACUCGGCAUU .......--------------..((((((((((...((((((((....))).....((((...(((...--(((((.....)))))..)))..)))))))))....)))...))))))). ( -25.30) >DroEre_CAF1 153678 113 - 1 ----G-AACAUUGCGAGGGAAUCAUCCCGAAUCACCUUGCUAACUGAUGUUUCAAUUCGGAAUGCAAAC--UCUACUGGUAGUAAAGCAGCUCCAAUCGCAAACCCGAUUACUCGGCAUU ----.-....((((((((((....))))........((((...((((.........))))...))))..--.....(((.(((......)))))).))))))..((((....)))).... ( -26.70) >DroYak_CAF1 141656 118 - 1 AACAAAAUCAUAGUAAGGCAUCCCUCUCGAAUCACCUUGCUAACUAAUGUUUCAAUUUGGAAUGCAAAU--UCCACAGAAAGUGAAGCUUCUCCAUGAGCAAACCCGAUUACUUGGCAUU ..(((((((.((((((((..((......))....))))))))......((((((...((((((....))--)))).......))))))..................))))..)))..... ( -21.30) >consensus AACAAAA______________CCAUGCCGAAUCACCUUGCUAACUAAUGUUUCCAUUUGGAAUGCAAAG__UCUACUGAAAGUAGAGCUGCUCCCAAAGCAAACCCGAUUACUCGGCAUU .......................((((((((((...((((...((((.........))))...))))....(((((.....)))))....................)))...))))))). (-14.40 = -14.56 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:37:23 2006