
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,268,677 – 25,268,811 |
| Length | 134 |
| Max. P | 0.718415 |

| Location | 25,268,677 – 25,268,786 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -13.27 |
| Energy contribution | -12.97 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
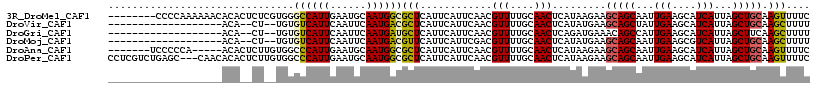
>3R_DroMel_CAF1 25268677 109 - 27905053 --------CCCCAAAAAACACACUCUCGUGGGCCAUUGAAUGCAAUGGCGCUCAUUCAUUCAACGUUUUGCAACUCAUAAGAAGCAGCAAUUGAAGCAUCAUUAGCUGCAAGUUUUC --------......(((((....(((..((((((((((....)))))))((..................))...)))..))).(((((...(((....)))...)))))..))))). ( -27.17) >DroVir_CAF1 176530 94 - 1 -------------------ACA--CU--UGUGUCAUUCAAUUCAAUGACGCUCAUUCAUUCAACGUUUUGCAACUCAUAUGAAGCAGCUAUUGAAGCAUCAUUAGCUGCAAGCUUUU -------------------...--..--.((((((((......))))))))...(((((.....(((....)))....)))))(((((((.(((....))).)))))))........ ( -24.60) >DroGri_CAF1 253896 94 - 1 -------------------ACA--CU--UGUGUCAUUCAAUUCAAUGAUGCUCAUUCAUUCAACGUUUUGCAACUCAGAUGAAACAGCCAUUGAAGCAUCAUUAGCUUCAAGCUUUU -------------------...--..--.((((((((......))))))))...((((((....(((....)))...))))))..(((..(((((((.......))))))))))... ( -18.10) >DroMoj_CAF1 163087 94 - 1 -------------------ACA--CU--UGUGUCAUUCAAUUCAAUGACGUUCAUUCAUUCGACGUUUUGCAACUCAUAUGAAGCAGCAAUUGAAGCGUCAUUAGCUGCAAGCUUUU -------------------...--..--...((((((......)))))).....(((((..((.(((....)))))..)))))(((((...(((....)))...)))))........ ( -19.10) >DroAna_CAF1 121808 105 - 1 -------UCCCCCA-----ACACUCUUGUGGCCCAUUGAAUGCAAUGGCGCUCAUUCAUUCAACGUUUUGCAACUCAUAAGAAGCAGCAAUUGAAGCAUCAUUAGCUGCAAGUUUUC -------......(-----((..(((((((((((((((....)))))).)).............(((....))).))))))).(((((...(((....)))...)))))..)))... ( -25.90) >DroPer_CAF1 130157 114 - 1 CCUCGUCUGAGC---CAACACACUCUUGUGGCCCAUUGAAUGCAAUGGCGCUCAUUCAUUCAACGUUUUGCAACUCAUAAGAAGCAGCAAUUGAAGCAUCAUUAGCUGCAAGUUUUC ....(..(((((---...((((....))))(((.((((....))))))))))))..)..........................(((((...(((....)))...)))))........ ( -27.10) >consensus ___________________ACA__CU__UGGGCCAUUCAAUGCAAUGACGCUCAUUCAUUCAACGUUUUGCAACUCAUAAGAAGCAGCAAUUGAAGCAUCAUUAGCUGCAAGCUUUC ...............................((((((......))))))(((............(((....))).........(((((...(((....)))...))))).))).... (-13.27 = -12.97 + -0.30)


| Location | 25,268,715 – 25,268,811 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.20 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -20.95 |
| Energy contribution | -21.97 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25268715 96 + 27905053 UAUGAGUUGCAAAACGUUGAAUGAAUGAGCGCCAUUGCAUUCAAUGGCCCACGAGAGUGUGUUUUUUGGGG----------------------GAGCAGGGGCCCAAGGGAUCCCAGG .......(((....(((((((((((((.....)))).))))))))).(((.((((((.....)))))))))----------------------..)))((((((....)).))))... ( -33.60) >DroPse_CAF1 129126 94 + 1 UAUGAGUUGCAAAACGUUGAAUGAAUGAGCGCCAUUGCAUUCAAUGGGCCACAAGAGUGUGUUG---GCUCAGACGAGGGCG--G------------------CGGAGGGGU-GCAAG ......(((((...(((((((((((((.....)))).))))))))).((((((....)))....---((((......)))))--)------------------).......)-)))). ( -27.70) >DroSec_CAF1 125683 106 + 1 UAUGAGUUGCAAAACGUUGAAUGAAUGAGCGCCAUUGCAUUCAAUGGCCCACGAGAGUCUGUUUUUCGGGGAGCGGGGGU------------GCAGCAGGGAGUCAAGGGAUCCCAGG .....(((((....(((((((((((((.....)))).)))))))))((((((....))(((((((....)))))))))))------------))))).((((.((....))))))... ( -39.00) >DroSim_CAF1 128107 106 + 1 UAUGAGUUGCAAAACGUUGAAUGAAUGAGCGCCAUUGCAUUCAAUGGCCCACGAGAGUGUGUUUUUCGGGGAGCGGAGGU------------GCAGCAGGGAGCCAAGGGAUCCCAGG .....((((((...(((((((((((((.....)))).)))))))))((((.((((((.....)))))).)).)).....)------------))))).((((.((...)).))))... ( -38.90) >DroEre_CAF1 142986 118 + 1 UAUGAGUUGCAAAACGUUGAAUGAAUGAGCGCCAUUGCAUUCAAUGGCCCACGAGAGUGUGUUUUUCGCGCAGUGGAGGGAGCAGGGAGCAGGGAGCAGGGAGCCCAGGGAUCCCAGG .......(((....(((((((((((((.....)))).))))))))).(((.(.(..(((((.....)))))..).).))).)))((((.(..((.((.....))))..)..))))... ( -39.00) >DroYak_CAF1 130695 114 + 1 UAUGAGUUGCAAAACGUUGAAUGAAUGAGCGCCAUUGCAUUCAAUGGCCCACGAGAGUGUGUUUUUCGAGCAGUGGAGGGAGCGGGGAGUCAAG----GGAAUCCCAGGGAUCCCAGG ......((((....(((((((((((((.....)))).)))))))))((.(((....))).)).......))))....((((...((((.((...----.)).)))).....))))... ( -35.90) >consensus UAUGAGUUGCAAAACGUUGAAUGAAUGAGCGCCAUUGCAUUCAAUGGCCCACGAGAGUGUGUUUUUCGGGCAGCGGAGGG____________GGAGCAGGGAGCCAAGGGAUCCCAGG ..((((..(((...(((((((((((((.....)))).)))))))))...(((....))))))..))))..............................((((.(.....).))))... (-20.95 = -21.97 + 1.02)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:37:18 2006