
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,251,196 – 25,251,336 |
| Length | 140 |
| Max. P | 0.906269 |

| Location | 25,251,196 – 25,251,300 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -5.51 |
| Energy contribution | -5.35 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.25 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
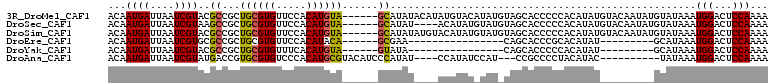
>3R_DroMel_CAF1 25251196 104 + 27905053 ACAAUGAUUAAUCGUACGCCGCUGCGUGUUCCACAUGUA------GCAUAUACAUAUGUACAUAUGUAGCACCCCCACAUAUGUACAAUAUGUAUAAAUGGACUCCAAAA ...((((....))))...((((((((((.....))))))------)).(((((((((((((((((((.(.....).)))))))))).)))))))))...))......... ( -34.30) >DroSec_CAF1 108496 100 + 1 ACAAUGAUUAAUCGUAAGCCGCUGCGUGUUCCACAUGUA------GCAUAU----ACAUAUGUAUGUAGCACCCCCACAUAUGUACAAUAUGUAUAAAUGGACUCCAAAA ...((((....)))).((((((((((((.....))))))------)).(((----(((((((((((((...........))))))).)))))))))...)).))...... ( -26.80) >DroSim_CAF1 110640 104 + 1 ACAAUGAUUAAUCGUACGCCGCUGCGUGUUCCACAUGUA------GCAUAUAUGUACAUAUGUAUGUAGCACCCCCACAUAUGUACAAUAUGUAUAAAUGGACUCCAAAA ...((((....))))...((((((((((.....))))))------))((((((((((((((((.............)))))))))).))))))......))......... ( -28.12) >DroEre_CAF1 125303 79 + 1 ACAAUGAUUAAUCGUGCGCCGCUGCGUGUUCCACAUACA------GCGAA----------------CAGCACCCGCACAUAU---------GCAUAAAUGGACUCCAAAA .............((((..(((((..((.....))..))------)))..----------------..))))..(((....)---------)).....(((...)))... ( -18.10) >DroYak_CAF1 112733 79 + 1 ACAAUGAUUAAUCGUACGCCGCUGCGUGUUUCACAUGUA------GUAUA----------------CAGCACCCCCACAUAU---------GCAUAAAUGGACUCCAAAA ...((((....))))...((((((((((.....))))))------))...----------------..(((..........)---------))......))......... ( -12.90) >DroAna_CAF1 104570 93 + 1 ACAAUGAUUAAUCGUAUGACCGUGCGUGUCCCACAUGCGUACAUCCCAUAU----CCAUAUCCAU---CCGCCCCUACAUAC----------UAUAAAUGGACUCCAAAA .............(((((...((((((((.....))))))))....)))))----.....(((((---..............----------.....)))))........ ( -11.51) >consensus ACAAUGAUUAAUCGUACGCCGCUGCGUGUUCCACAUGUA______GCAUAU____ACAUAU_UAU_UAGCACCCCCACAUAU_________GUAUAAAUGGACUCCAAAA ...((((....))))..((...((((((.....))))))......))...................................................(((...)))... ( -5.51 = -5.35 + -0.16)

| Location | 25,251,227 – 25,251,336 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -16.43 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25251227 109 - 27905053 CUCGCGCUCUUAAAGCCUACGGAAAGUUUUGUGACUUUUUGGAGUCCAUUUAUACAUAUUGUACAUAUGUGGGGGUGCUACAUAUGUACAUAUGUAUAUGC------UACAUGUG (((..(((.....)))...(((((((((....))))))))))))......(((((((((.((((((((((((.....)))))))))))))))))))))...------........ ( -35.60) >DroSec_CAF1 108527 105 - 1 CUCGCGCUCUUAAAGCCUACGGAAAGUUUUGUGACUUUUUGGAGUCCAUUUAUACAUAUUGUACAUAUGUGGGGGUGCUACAUACAUAUGU----AUAUGC------UACAUGUG ...(((((((......((.(((((((((....))))))))).))........(((((((.....))))))))))))))......(((((((----(.....------)))))))) ( -27.90) >DroSim_CAF1 110671 109 - 1 CUCGCGCUCUUAAAGCCUACGGAAAGUUUUGUGACUUUUUGGAGUCCAUUUAUACAUAUUGUACAUAUGUGGGGGUGCUACAUACAUAUGUACAUAUAUGC------UACAUGUG (((..(((.....)))...(((((((((....))))))))))))..........(((((((((((((((((..(......).)))))))))))).))))).------........ ( -29.10) >DroEre_CAF1 125334 84 - 1 CUCGCGCUCUUAAAGCCUACGGAAAGUUUUGUGACUUUUUGGAGUCCAUUUAUGC---------AUAUGUGCGGGUGCUG----------------UUCGC------UGUAUGUG (((..(((.....)))...(((((((((....)))))))))))).........((---------(((((.(((..(...)----------------..)))------))))))). ( -20.50) >DroYak_CAF1 112764 84 - 1 CUCGCGCUCUUAAAGCCUAUGGAAAGUUUUGUGACUUUUUGGAGUCCAUUUAUGC---------AUAUGUGGGGGUGCUG----------------UAUAC------UACAUGUG ...(((((((....((.((((.(.((...((.(((((....))))))).)).).)---------))).)).)))))))..----------------.....------........ ( -18.60) >DroAna_CAF1 104601 98 - 1 CUCGCGCUCUUAAAGCCUACGGAAAGUUUUGUGACUUUUUGGAGUCCAUUUAUA----------GUAUGUAGGGGCGG---AUGGAUAUGG----AUAUGGGAUGUACGCAUGUG ...(((.(((((...((..(((((((((....)))))))))..(((((((....----------((........)).)---))))))..))----...)))))....)))..... ( -22.40) >consensus CUCGCGCUCUUAAAGCCUACGGAAAGUUUUGUGACUUUUUGGAGUCCAUUUAUAC_________AUAUGUGGGGGUGCUA_AUA_AUAUGU____AUAUGC______UACAUGUG ...((((((.......((.(((((((((....))))))))).)).((((.(((...........))).))))))))))..................................... (-16.43 = -16.52 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:37:06 2006