
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,247,275 – 25,247,397 |
| Length | 122 |
| Max. P | 0.718089 |

| Location | 25,247,275 – 25,247,365 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -15.63 |
| Energy contribution | -16.66 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25247275 90 + 27905053 CGCAGAGCUUAUACGCAGCAUAGCAUACUUU-UUUGGGCAGCGUGUUCUCAAAAUGAAGCUGCCCUACCCGCCGCCAUCACCAUCUCCAUU .((.(.((......((......)).......-...(((((((...(((.......)))))))))).....)))))................ ( -18.90) >DroSec_CAF1 104536 90 + 1 CGCACAGCUUAUACGCAGCAUAGCAUACUUU-UUUGGGCAGCGCGUUCUCAAAAUGAAGCUGCCCUGCCCGCUGCCAUCUCCAUCUCCAUU ..............(((((...(((......-...(((((((.((((.....))))..))))))))))..)))))................ ( -24.20) >DroSim_CAF1 106677 90 + 1 CGCACAGCUUAUACGCAGCAUAGCAUACUUU-UUUGGGCAGCGCGUUCUCAAAAUGAAGCUGCCCUGCCCGCUGCCAUCUCCAUCUCCAUU ..............(((((...(((......-...(((((((.((((.....))))..))))))))))..)))))................ ( -24.20) >DroEre_CAF1 121306 84 + 1 UGCACAGCUUACACGCAGCAUAGCAUACUUU-UUUGGGCAGAGCGUUCCCAAAAUGAAGCUGCCCUGUCCGCUGCCAUCUCCAUC------ ....((((..(((.((......)).......-...((((((..((((.....))))...)))))))))..))))...........------ ( -21.60) >DroYak_CAF1 108796 83 + 1 --CACAGCUUACACGCAGCAUAGCAUACUUUUUUUGGGCAGCGCGUUCCCAAAAUUAAGCUGCCCUGCCCGCUGCCAUCUCCAUC------ --............(((((...(((..........(((((((................))))))))))..)))))..........------ ( -22.69) >DroAna_CAF1 100478 72 + 1 ---GGGGCUUAUACGCGGUGUAGCAUUCUUU-UUUGGGCAGCGCGUACACAAAAUGAAGCUGUCC----------CAUCUCCAUCG----- ---(((((...((((((.(((..((......-..)).))).))))))((.....)).....))))----------)..........----- ( -20.70) >consensus CGCACAGCUUAUACGCAGCAUAGCAUACUUU_UUUGGGCAGCGCGUUCUCAAAAUGAAGCUGCCCUGCCCGCUGCCAUCUCCAUCU_____ ....((((......((......))...........(((((((................))))))).....))))................. (-15.63 = -16.66 + 1.03)

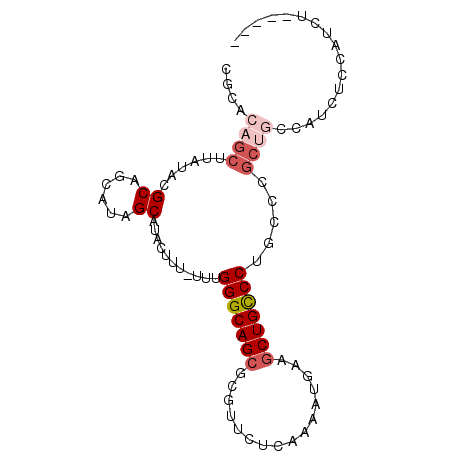

| Location | 25,247,275 – 25,247,365 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -20.13 |
| Energy contribution | -21.36 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25247275 90 - 27905053 AAUGGAGAUGGUGAUGGCGGCGGGUAGGGCAGCUUCAUUUUGAGAACACGCUGCCCAAA-AAAGUAUGCUAUGCUGCGUAUAAGCUCUGCG ...((((...(((...((((((....((((((((((.......)))...)))))))...-..((....)).)))))).)))...))))... ( -28.20) >DroSec_CAF1 104536 90 - 1 AAUGGAGAUGGAGAUGGCAGCGGGCAGGGCAGCUUCAUUUUGAGAACGCGCUGCCCAAA-AAAGUAUGCUAUGCUGCGUAUAAGCUGUGCG ................(((((((((((.((..(((......)))...)).))))))...-...((((((......))))))..)))))... ( -32.10) >DroSim_CAF1 106677 90 - 1 AAUGGAGAUGGAGAUGGCAGCGGGCAGGGCAGCUUCAUUUUGAGAACGCGCUGCCCAAA-AAAGUAUGCUAUGCUGCGUAUAAGCUGUGCG ................(((((((((((.((..(((......)))...)).))))))...-...((((((......))))))..)))))... ( -32.10) >DroEre_CAF1 121306 84 - 1 ------GAUGGAGAUGGCAGCGGACAGGGCAGCUUCAUUUUGGGAACGCUCUGCCCAAA-AAAGUAUGCUAUGCUGCGUGUAAGCUGUGCA ------..........(((((..(((.(.((((....(((((((.........))))))-).((....))..))))).)))..)))))... ( -25.90) >DroYak_CAF1 108796 83 - 1 ------GAUGGAGAUGGCAGCGGGCAGGGCAGCUUAAUUUUGGGAACGCGCUGCCCAAAAAAAGUAUGCUAUGCUGCGUGUAAGCUGUG-- ------..........(((((((((((.((..((((....))))...)).))))))........(((((......)))))...))))).-- ( -30.50) >DroAna_CAF1 100478 72 - 1 -----CGAUGGAGAUG----------GGACAGCUUCAUUUUGUGUACGCGCUGCCCAAA-AAAGAAUGCUACACCGCGUAUAAGCCCC--- -----.(((((((.((----------...)).)))))))...((((((((.((..((..-......))...)).))))))))......--- ( -14.70) >consensus _____AGAUGGAGAUGGCAGCGGGCAGGGCAGCUUCAUUUUGAGAACGCGCUGCCCAAA_AAAGUAUGCUAUGCUGCGUAUAAGCUGUGCG ................(((((.....(((((((................))))))).......((((((......))))))..)))))... (-20.13 = -21.36 + 1.22)



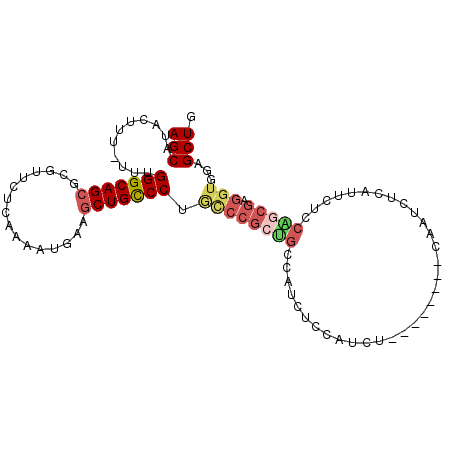
| Location | 25,247,296 – 25,247,397 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.77 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25247296 101 + 27905053 AGCAUACUUU-UUUGGGCAGCGUGUUCUCAAAAUGAAGCUGCCCUACCCGCCGCCAUCACCAUCUCCAUUCUCCAAUCUCAUUCUCCGGCGAGGUAGAGCUG (((.......-...(((((((...(((.......))))))))))(((((((((.................................))))).))))..))). ( -28.61) >DroSec_CAF1 104557 101 + 1 AGCAUACUUU-UUUGGGCAGCGCGUUCUCAAAAUGAAGCUGCCCUGCCCGCUGCCAUCUCCAUCUCCAUUCUCCAAUCUCAUUCUCCAGCGAGGUGGAGCUG .(((......-...(((((((.((((.....))))..))))))).......)))((.((((((((((.....................).))))))))).)) ( -28.99) >DroSim_CAF1 106698 101 + 1 AGCAUACUUU-UUUGGGCAGCGCGUUCUCAAAAUGAAGCUGCCCUGCCCGCUGCCAUCUCCAUCUCCAUUCUCCAAUCUCGUUCUCCGGCGAGGUGGAGCUG (((.......-...((((((.(((((.((.....))))).)).))))))......................((((.((((((......))))))))))))). ( -31.10) >DroEre_CAF1 121327 87 + 1 AGCAUACUUU-UUUGGGCAGAGCGUUCCCAAAAUGAAGCUGCCCUGUCCGCUGCCAUCUCCAUC--------------UGAUUCUCCAGCGAGGUGAGGCUG ..........-...((((((..((((.....))))...)))))).....(((..((((((...(--------------((......))).)))))).))).. ( -26.30) >DroYak_CAF1 108815 88 + 1 AGCAUACUUUUUUUGGGCAGCGCGUUCCCAAAAUUAAGCUGCCCUGCCCGCUGCCAUCUCCAUC--------------UGAUUCUCCACCGAGGUGAAGCUG ..............((((((.(((((..........))).)).))))))(((..((((((....--------------((......))..)))))).))).. ( -23.80) >DroAna_CAF1 100496 83 + 1 AGCAUUCUUU-UUUGGGCAGCGCGUACACAAAAUGAAGCUGUCC----------CAUCUCCAUCG--------CGAUCUCCGUAUCCAACGACGUGGAGCUA .((.......-..((((((((.(((.......)))..)))).))----------))........)--------)...(((((..((....))..)))))... ( -20.63) >consensus AGCAUACUUU_UUUGGGCAGCGCGUUCUCAAAAUGAAGCUGCCCUGCCCGCUGCCAUCUCCAUCU________CAAUCUCAUUCUCCAGCGAGGUGGAGCUG (((...........(((((((................))))))).((((((((.................................))))).)))...))). (-17.74 = -18.77 + 1.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:37:04 2006