
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
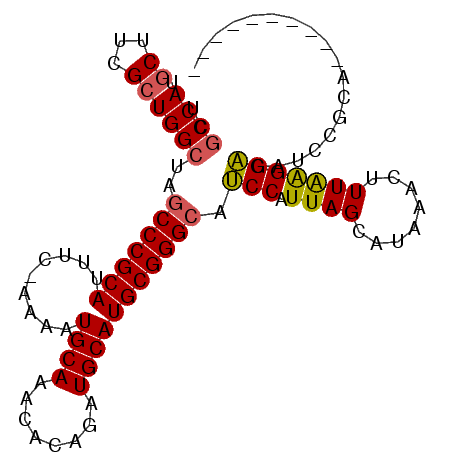
| Location | 25,234,926 – 25,235,016 |
| Length | 90 |
| Max. P | 0.870480 |

| Location | 25,234,926 – 25,235,016 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -21.33 |
| Energy contribution | -21.23 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25234926 90 + 27905053 UCGCCAUGCUUCGCUGGCUAGCCCGCAUUUU-AAAAUGCAAACACAGAUGCAUGCGGGCAUCCAUUAGCAUAAACUUUAAGGACAUUCGCA---------- ..((((.((...))))))..(((((((....-....((((........))))))))))).(((.((((........)))))))........---------- ( -24.40) >DroSec_CAF1 92035 90 + 1 UCGCCAUGCUUCGCUGGCUAGCCCGCAUUUC-AAAAUGCAAACACAGAUGCAUGCGGGCAUCCAUUAGCAUAAACUUUAAGGACAUCCGCA---------- ..((.(((((....(((...(((((((....-....((((........)))))))))))..)))..))))).........((....)))).---------- ( -26.70) >DroSim_CAF1 93690 90 + 1 UCGCCAUGCUUCGCUGGCUAGCCCGCAUUUC-AAAAUGCAAACACAGAUGCAUGCGGGCAUCCAUUAGCAUAAACUUUAAGGACAUCCGCA---------- ..((.(((((....(((...(((((((....-....((((........)))))))))))..)))..))))).........((....)))).---------- ( -26.70) >DroEre_CAF1 100607 90 + 1 UCGCCAUGCCUCGCUGGCUAGCCCGCAUUUC-AAAAUGCAAACCCAGAUGCAUGCGGGCAUCCAUUAGCAUAAACUUUGGGGACAUCCGCA---------- ..((((.((...))))))..(((((((....-....((((........)))))))))))..(((..((......)).)))((....))...---------- ( -26.10) >DroYak_CAF1 96060 90 + 1 UCGCCAUGCUUCGCUGGCUAGCCCGCAUCUC-AAAAUGCAAACCAAGAUGCAUGCGGGCAUCCAUUAGCAUAAACUUUAGGGACAUCCGCA---------- ..((.(((((....(((...(((((((....-....((((........)))))))))))..)))..))))).........((....)))).---------- ( -26.50) >DroAna_CAF1 88949 94 + 1 UCCCCA-------CUGGUUAACCCGCAUGACUCGAAUGCAAAUACAAGUGCAUGCGGGCGGCCAUUAGCAUCAACUUUAAGGUCCUUUACACACACAGUCA .....(-------(((.....(((((((((((.(.((....)).).))).)))))))).((((.((((........))))))))...........)))).. ( -23.80) >consensus UCGCCAUGCUUCGCUGGCUAGCCCGCAUUUC_AAAAUGCAAACACAGAUGCAUGCGGGCAUCCAUUAGCAUAAACUUUAAGGACAUCCGCA__________ ..((((.((...))))))..(((((((.........((((........))))))))))).(((.((((........))))))).................. (-21.33 = -21.23 + -0.09)


| Location | 25,234,926 – 25,235,016 |
|---|---|
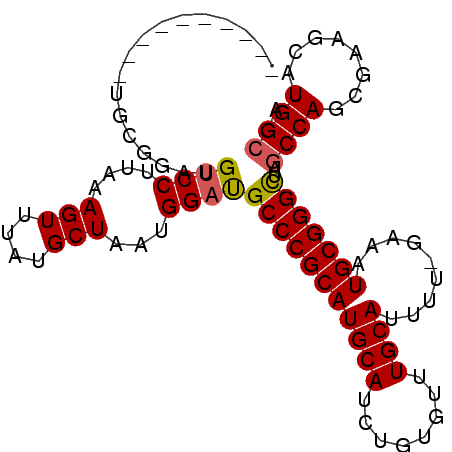
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -24.83 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25234926 90 - 27905053 ----------UGCGAAUGUCCUUAAAGUUUAUGCUAAUGGAUGCCCGCAUGCAUCUGUGUUUGCAUUUU-AAAAUGCGGGCUAGCCAGCGAAGCAUGGCGA ----------.......((((....(((....)))...))))((((((((......(((....)))...-...))))))))..((((((...)).)))).. ( -29.30) >DroSec_CAF1 92035 90 - 1 ----------UGCGGAUGUCCUUAAAGUUUAUGCUAAUGGAUGCCCGCAUGCAUCUGUGUUUGCAUUUU-GAAAUGCGGGCUAGCCAGCGAAGCAUGGCGA ----------.((((....))........((((((..(((..((((((((......(((....)))...-...))))))))...)))....)))))))).. ( -32.10) >DroSim_CAF1 93690 90 - 1 ----------UGCGGAUGUCCUUAAAGUUUAUGCUAAUGGAUGCCCGCAUGCAUCUGUGUUUGCAUUUU-GAAAUGCGGGCUAGCCAGCGAAGCAUGGCGA ----------.((((....))........((((((..(((..((((((((......(((....)))...-...))))))))...)))....)))))))).. ( -32.10) >DroEre_CAF1 100607 90 - 1 ----------UGCGGAUGUCCCCAAAGUUUAUGCUAAUGGAUGCCCGCAUGCAUCUGGGUUUGCAUUUU-GAAAUGCGGGCUAGCCAGCGAGGCAUGGCGA ----------(((((..((((....(((....)))...))))..)))))(((.....((((((((((..-..)))))))))).(((.....)))...))). ( -31.30) >DroYak_CAF1 96060 90 - 1 ----------UGCGGAUGUCCCUAAAGUUUAUGCUAAUGGAUGCCCGCAUGCAUCUUGGUUUGCAUUUU-GAGAUGCGGGCUAGCCAGCGAAGCAUGGCGA ----------(((((..((((....(((....)))...))))..)))))(((((((..(........).-.))))))).....((((((...)).)))).. ( -32.70) >DroAna_CAF1 88949 94 - 1 UGACUGUGUGUGUAAAGGACCUUAAAGUUGAUGCUAAUGGCCGCCCGCAUGCACUUGUAUUUGCAUUCGAGUCAUGCGGGUUAACCAG-------UGGGGA (.((((.((.......((.((....(((....)))...))))(((((((((.(((((..........))))))))))))))..)))))-------).)... ( -30.60) >consensus __________UGCGGAUGUCCUUAAAGUUUAUGCUAAUGGAUGCCCGCAUGCAUCUGUGUUUGCAUUUU_GAAAUGCGGGCUAGCCAGCGAAGCAUGGCGA .................((((....(((....)))...))))(((((((((((........)))).........)))))))..((((........)))).. (-24.83 = -24.88 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:36:58 2006