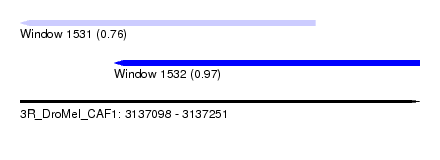
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,137,098 – 3,137,251 |
| Length | 153 |
| Max. P | 0.971063 |

| Location | 3,137,098 – 3,137,211 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -13.69 |
| Energy contribution | -13.17 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
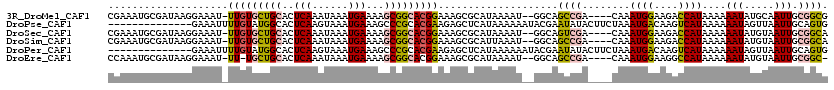
>3R_DroMel_CAF1 3137098 113 - 27905053 CGAAAUGCGAUAAGGAAAU-UUGUGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUAAAAU--GGCAGCCGA----CAAAUGGAAGACCAUAAAAAAUAUGCAAUUGCGGCG ((...((((((.......(-(((((((((..(((......)))...))))))))))....(((((..((--(((..(((.----....)))..).))))......))))).)))))).)) ( -29.30) >DroPse_CAF1 11571 106 - 1 --------------GAAAUUUUGUAUGGCACUCAAGUAAAUGAAAGCCCGCACGAAGAGCUCAUAAAAAAUACGAAUAUACUUCUAAAUGACAAGUCAUAAAAAAUAGUUAAUUGCAGUG --------------.....((((((((((..(((......)))..))).((.......)).........)))))))...........((((....))))..................... ( -11.90) >DroSec_CAF1 13022 113 - 1 CGAAAUGCGAUAAGGAAAU-UUGUGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUAAAAU--GGCAGUCGA----CAAAUGGAAGACCAUAAAAAAUAUGUAAUUGCGGCA .....(((..........(-(((((((((..(((......)))...))))))))))...((((......--.(((.....----...((((....))))........)))...))))))) ( -25.79) >DroSim_CAF1 13533 113 - 1 CGAAAUGCGAUAAGGAAAU-UUGUGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUUAAAU--GGCAGCCGA----CAAAUGGAAGACCAUAAAAAAUAUGUAAUUGCGGCA ...((((((.........(-(((((((((..(((......)))...))))))))))...))))))....--....((((.----(((((((....)))).............))))))). ( -28.71) >DroPer_CAF1 11544 106 - 1 --------------GAAAUUUUGUAUGGCACUCAAGUAAAUGAAAGCCCGCACGAAGAGCUCAUAAAAAAUACGAAUAUACUUCUAAAUGACAAGUCAUAAAAAAUAGUUAAUUGCAGUG --------------.....((((((((((..(((......)))..))).((.......)).........)))))))...........((((....))))..................... ( -11.90) >DroEre_CAF1 13499 111 - 1 CCAAAUGCGAUAAGGAAAU-UU-UGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUAAAAU--GGCAGCCGA----CAAAUGGAAGGCCAUAAAAAAUAUGUAAUUGCGGC- .....((((((........-..-((((((..(((......)))...))))))........(((((..((--(((..(((.----....)))...)))))......))))).))))))..- ( -24.60) >consensus CGAAAUGCGAUAAGGAAAU_UUGUGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUAAAAU__GGCAGCCGA____CAAAUGGAAGACCAUAAAAAAUAUGUAAUUGCGGCG ....................(((((((((..(((......)))...)))))))))....................((((........((((....))))....(((.....))).)))). (-13.69 = -13.17 + -0.52)
| Location | 3,137,134 – 3,137,251 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -16.56 |
| Energy contribution | -16.45 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
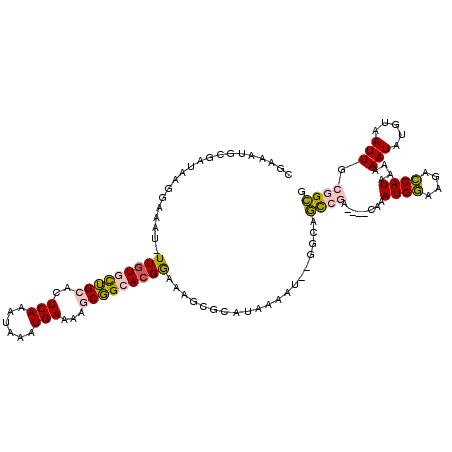
>3R_DroMel_CAF1 3137134 117 - 27905053 AAGUGGCAUUGAUGACAGUGCAAACAAGAGCGGCGACAGUCGAAAUGCGAUAAGGAAAU-UUGUGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUAAAAU--GGCAGCCGA .....((((((....)))))).........((((....((((..(((((.........(-(((((((((..(((......)))...))))))))))...)))))....)--))).)))). ( -33.90) >DroPse_CAF1 11611 94 - 1 AAAUGGCAUUGAUGACAGUGCAAACAAG--------------------------GAAAUUUUGUAUGGCACUCAAGUAAAUGAAAGCCCGCACGAAGAGCUCAUAAAAAAUACGAAUAUA .....((((((....)))))).......--------------------------.....((((((((((..(((......)))..))).((.......)).........))))))).... ( -16.70) >DroSec_CAF1 13058 117 - 1 AAGUGGCAUUGAUGACAGUGCAAACAAGAGCGGCGACAGUCGAAAUGCGAUAAGGAAAU-UUGUGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUAAAAU--GGCAGUCGA .....((((((....))))))............((((.((((..(((((.........(-(((((((((..(((......)))...))))))))))...)))))....)--))).)))). ( -32.90) >DroSim_CAF1 13569 117 - 1 AAGUGGCAUUGAUGACAGUGCAAACAAGAGCGGCGACAGUCGAAAUGCGAUAAGGAAAU-UUGUGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUUAAAU--GGCAGCCGA .....((((((....)))))).........((((....((((.((((((.........(-(((((((((..(((......)))...))))))))))...))))))...)--))).)))). ( -35.10) >DroPer_CAF1 11584 94 - 1 AAAUGGCAUUGAUGACAGUGCAAACAAG--------------------------GAAAUUUUGUAUGGCACUCAAGUAAAUGAAAGCCCGCACGAAGAGCUCAUAAAAAAUACGAAUAUA .....((((((....)))))).......--------------------------.....((((((((((..(((......)))..))).((.......)).........))))))).... ( -16.70) >DroEre_CAF1 13534 116 - 1 AAGUGCCAUUGAUGACAGUGCAAACAAGAGCGGCGACAGUCCAAAUGCGAUAAGGAAAU-UU-UGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUAAAAU--GGCAGCCGA ..(((((((((....))))((........(((((((...(((...........)))...-.)-))))))..(((......)))...)))))))((...((.(((...))--)))..)).. ( -29.10) >consensus AAGUGGCAUUGAUGACAGUGCAAACAAGAGCGGCGACAGUCGAAAUGCGAUAAGGAAAU_UUGUGCUGCACUCAAAUAAAUGAAAAGCGGCACGGAAAGCGCAUAAAAU__GGCAGCCGA ..(((((((((....)))))).......................................(((((((((..(((......)))...)))))))))......)))................ (-16.56 = -16.45 + -0.11)

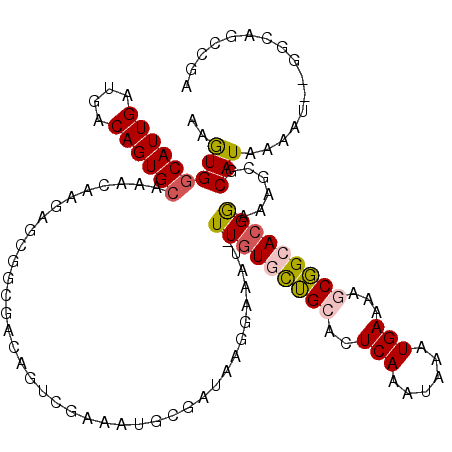
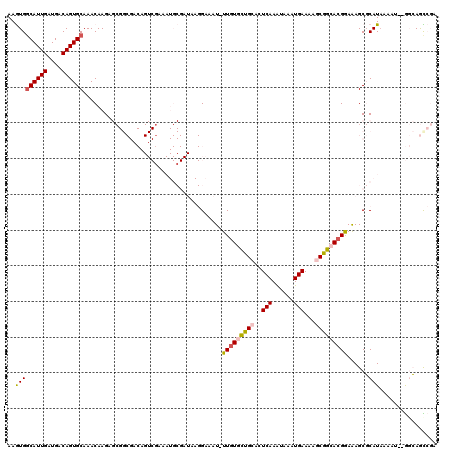
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:35 2006