| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
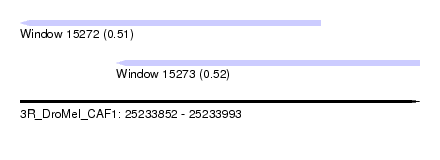
| Location | 25,233,852 – 25,233,993 |
| Length | 141 |
| Max. P | 0.522190 |

| Location | 25,233,852 – 25,233,958 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -11.88 |
| Energy contribution | -11.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
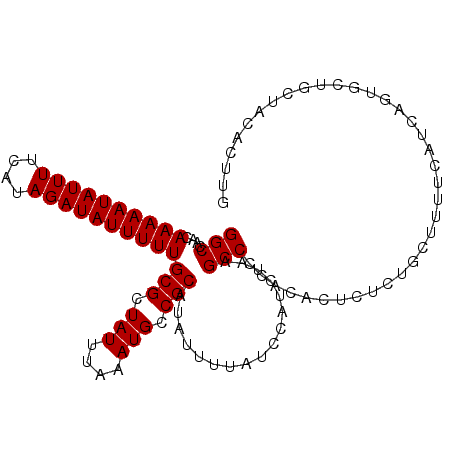
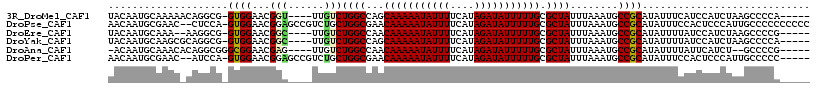
>3R_DroMel_CAF1 25233852 106 - 27905053 GGCCAGCAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUCAUCCAUCUAAGCCCCAAACUCUCGGCUUUUCAUCAGAGCUGCUACACUUG (((..((((((((((((....))))))))))))((.........))....................))).........(((((((.....)))))))......... ( -21.50) >DroSim_CAF1 92635 106 - 1 GGCCAGCAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUUAUCCAUCUAAGCCCCAAACUCUCGGCUUUUCAUCAGAGCUGCUACACUGA (((..((((((((((((....))))))))))))((.........))....................))).........(((((((.....)))))))......... ( -21.50) >DroEre_CAF1 93933 106 - 1 GGCCAACAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUUAUCCAUCUAAGCCCCGCACUCUCUGCCUUUCAUUAGUGCUGCUACACUUG (((....((((((((((....))))))))))(((.(((....))).))).................)))..(((((...((.....))..)))))........... ( -16.40) >DroYak_CAF1 94968 106 - 1 GGCCAGCAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUUAUCCAUCUAAGCCCCACACUCCCUGCUUUUCAUCAGUGCUGCUACAUUUG (((..((((((((((((....)))))))))))))))...((((((..(((((((..........((((............))))......)))).)))..)))))) ( -17.99) >DroAna_CAF1 87936 104 - 1 GGCCAACAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUUAUUCAUCU--GCCCCGCACUUGCUCUUUGUCAAGAGUGUUUCCAAUUGUU (((....((((((((((....))))))))))(((.(((....))).)))...............--))).(((.(((((.....).)))).)))............ ( -17.10) >DroPer_CAF1 96840 97 - 1 GGCGAACAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUCCACUCCCAUUGCCCCC---CCGC-UCUUUCCAUCAGUCUAUCUCG----- (((((..((((((((((....))))))))))(((.(((....))).)))...............)))))...---....-.....................----- ( -14.10) >consensus GGCCAACAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUUAUCCAUCUAAGCCCCACACUCUCUGCUUUUCAUCAGUGCUGCUACACUUG (((....((((((((((....))))))))))(((.(((....))).))).................)))..................................... (-11.88 = -11.88 + 0.00)


| Location | 25,233,886 – 25,233,993 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.48 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25233886 107 - 27905053 UACAAUGCAAAAACAGGCG-GUGGAACGGU----UUGUCUGGCCAGCAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUCAUCCAUCUAAGCCCCA----- ...............((((-(((((.((((----(((..((((..((((((((((((....))))))))))))))))..)))..)))).........))))))...)))...----- ( -27.30) >DroPse_CAF1 95864 114 - 1 AACAAUGCGAAC--CUCCA-GUGGAACGGAGCCGUCUGCUGGCGAACAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUCCACUCCCAUUGCCCCCCCCCC ....(((((...--...((-(..((.((....))))..)))(((..(((((((((((....))))))))))))))...........))))).......................... ( -27.50) >DroEre_CAF1 93967 105 - 1 UACAAUGCAAA--AAGGCG-GUGGAACGGC----UUGUCUGGCCAACAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUUAUCCAUCUAAGCCCCG----- ...........--..((((-(((((.((((----(((..((((...(((((((((((....))))))))))).))))..)))..)))).........))))))...)))...----- ( -25.40) >DroYak_CAF1 95002 107 - 1 UACAAUGCAAGCGCAGGCG-GUGGAACGGC----UUGUCUGGCCAGCAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUUAUCCAUCUAAGCCCCA----- .....(((....)))((((-(((((..(((----(.....)))).((((((((((((....))))))))))))........................))))))...)))...----- ( -30.80) >DroAna_CAF1 87970 105 - 1 -ACAAUGCAAACACAGGCGGGCGGAACGAG----UUGUCUGGCCAACAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUUAUUCAUCU--GCCCCG----- -.....((........))(((((((..(((----(......((...(((((((((((....))))))))))).((.........)).)).......)))).)))--))))..----- ( -24.32) >DroPer_CAF1 96865 109 - 1 AACAAUGCGAAC--AUCCA-GUGGAACGGAGCCGUCUGCUGGCGAACAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUCCACUCCCAUUGCCCCC----- ....(((((...--...((-(..((.((....))))..)))(((..(((((((((((....))))))))))))))...........))))).....................----- ( -27.50) >consensus UACAAUGCAAAC_CAGGCG_GUGGAACGGA____UUGUCUGGCCAACAAAAAUAUUUUCAUAGAUAUUUUUGCGCUAUUUAAAUGCCGCAUAUUUCAUCCAUCUAAGCCCCA_____ ....................((((...(((......)))((((...(((((((((((....))))))))))).))))........))))............................ (-14.59 = -14.48 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:36:56 2006