
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,196,342 – 25,196,479 |
| Length | 137 |
| Max. P | 0.915419 |

| Location | 25,196,342 – 25,196,440 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.12 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -15.61 |
| Energy contribution | -17.05 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25196342 98 - 27905053 UCUCUGCAGAGAUGCAGCUAAA-------------CGCUGC----UUCCUUGGCUGAAUUCGUAUGCGA-----GGCAGCAUAUGGCAGCCGGAAAUGAAACGUAAAUCAAAUAAGUAAA ((((....)))).(((((....-------------.)))))----((((..(((((...((((((((..-----....)))))))))))))))))......................... ( -31.50) >DroPse_CAF1 59372 108 - 1 --------UAGUUGCAGCUAAAAUGCCUCUCUGUUCGCUGC----UUCCUUCGCUGAAUUCGUAUGCGAGGCGAGGCAGCAUAUGGCAGCCGGAAAUGAAACGUAAAUCAAAUAAGUAAG --------..(((((.((......))..........(((((----(((((((((...........)))))).)))))))).....))))).............................. ( -31.60) >DroWil_CAF1 136479 100 - 1 UCUCAA--------AAACUAAAACACCGCCAU---CGUCUC----UAACAUGACUGAAAUCGCUUGCCA-----GGCAGCAUAUGGCAGCCGGAAAUGAAACGUAAAUCAAAUAAGUAAG ......--------..(((......(((...(---((((..----......))).))....(((.((((-----.........))))))))))...(((........)))....)))... ( -14.90) >DroYak_CAF1 57432 98 - 1 UCUCUGCAGAUAUGCAGCUAAA-------------CGCUGC----UUCCUUGGCUGAAUUCGUAUGCGA-----GGCAGCAUAUGGCAGCCGGAAAUGAAACGUAAAUCAAAUAAGUAAA ....(((......(((((....-------------.)))))----((((..(((((...((((((((..-----....))))))))))))))))).......)))............... ( -29.70) >DroAna_CAF1 52721 103 - 1 AACCUGUACAAAUGCAGCUAAAA------------UGCCGCCGUGCUCCUUGGCUGAAUUCGUAUGCGA-----GGCAGCAUAUGGCUGCCGGAAAUGAAACGUAAAUCAAAUAAGUAAG ..((.((((.(((.(((((((..------------.((......))...))))))).))).))))....-----((((((.....))))))))........................... ( -27.30) >DroPer_CAF1 60332 108 - 1 --------UAGUUGCAGCUAAAAUGCCUCUCUGUUCGCUGC----UUCCUUCGCUGAAUUCGUAUGCGAGGCGAGGCAGCAUAUGGCAGCCGGAAAUGAAACGUAAAUCAAAUAAGUAAG --------..(((((.((......))..........(((((----(((((((((...........)))))).)))))))).....))))).............................. ( -31.60) >consensus UCUCUG__GAGAUGCAGCUAAAA____________CGCUGC____UUCCUUGGCUGAAUUCGUAUGCGA_____GGCAGCAUAUGGCAGCCGGAAAUGAAACGUAAAUCAAAUAAGUAAG .............(((((..................)))))....((((..(((((...((((((((...........)))))))))))))))))......................... (-15.61 = -17.05 + 1.45)



| Location | 25,196,382 – 25,196,479 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -23.41 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25196382 97 + 27905053 GCUGCCUCGCAUACGAAUUCAGCCAAGGAA----GCAGCG-UUUAGCUGCAUCUCUGCAGAGAGAGAAAAGUGGCAGCAAGUAUCGAGGAAGAUUCAUAAAA .((.(((((.(((((....).((((.....----(((((.-....))))).(((((......)))))....)))).....))))))))).)).......... ( -32.50) >DroSec_CAF1 55571 84 + 1 GCUGCCUCGCAUACGAAUUCAGCCAAGGAA----GCAGCG-UUUAGCUGCAUCUCUGCAGAGAGAGAAAAGUGGCAGCAAGUAUCGCGG------------- (((((((((....))).(((..........----(((((.-....))))).(((((....))))))))....))))))...........------------- ( -28.70) >DroSim_CAF1 54958 97 + 1 GCUGCCUCGCAUACGAAUUCAGCCAAGGAA----GCAGCG-UUUAGCUGCAUCUCUGCAGAGAGAGAAAAGCGGCAGCAAGUAUCGAGGGAGAUUCAUAAAU .((.(((((.(((((....).(((......----(((((.-....))))).(((((......))))).....))).....))))))))).)).......... ( -34.00) >DroEre_CAF1 58064 96 + 1 GCUGCCUCGCAUACGAAUUCAGCCAAGGAA----GCAGCG-UUUAGCUGCAUUUCUGCAGAGAGAGAAAAGUGGCGGCGAUGAUCGAG-GAGAUUCAUAAAU (((((((((....))).(((.((..(((((----(((((.-....))))).))))))).)))..........)))))).((((((...-..)).)))).... ( -29.40) >DroYak_CAF1 57472 97 + 1 GCUGCCUCGCAUACGAAUUCAGCCAAGGAA----GCAGCG-UUUAGCUGCAUAUCUGCAGAGAGAGAAAAGUGGCAGCGAUAAUCGAGGGAGAUUCAUAAGU .((.((((((....(....).((((.....----(((((.-....)))))...(((......)))......)))).))(.....))))).)).......... ( -25.80) >DroAna_CAF1 52761 84 + 1 GCUGCCUCGCAUACGAAUUCAGCCAAGGAGCACGGCGGCAUUUUAGCUGCAUUUGUACAGGUUCAGAAAAGUAGUACUCUGAAU------------------ ((((..(((....)))...))))....(.(((..(((((......)))))...))).)..(((((((..........)))))))------------------ ( -20.60) >consensus GCUGCCUCGCAUACGAAUUCAGCCAAGGAA____GCAGCG_UUUAGCUGCAUCUCUGCAGAGAGAGAAAAGUGGCAGCAAGUAUCGAGGGAGAUUCAUAAAU ((((((.(..........................(((((......))))).(((((......)))))...).))))))........................ (-23.41 = -23.08 + -0.33)
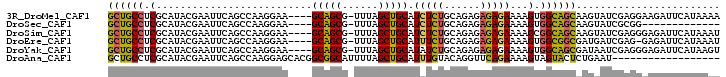


| Location | 25,196,382 – 25,196,479 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -17.59 |
| Energy contribution | -18.73 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25196382 97 - 27905053 UUUUAUGAAUCUUCCUCGAUACUUGCUGCCACUUUUCUCUCUCUGCAGAGAUGCAGCUAAA-CGCUGC----UUCCUUGGCUGAAUUCGUAUGCGAGGCAGC ..........((.(((((((((...(.((((....(((((......))))).(((((....-.)))))----.....)))).).....)))).))))).)). ( -28.70) >DroSec_CAF1 55571 84 - 1 -------------CCGCGAUACUUGCUGCCACUUUUCUCUCUCUGCAGAGAUGCAGCUAAA-CGCUGC----UUCCUUGGCUGAAUUCGUAUGCGAGGCAGC -------------...........((((((...........((.((.(((..(((((....-.)))))----...))).)).))..(((....))))))))) ( -26.80) >DroSim_CAF1 54958 97 - 1 AUUUAUGAAUCUCCCUCGAUACUUGCUGCCGCUUUUCUCUCUCUGCAGAGAUGCAGCUAAA-CGCUGC----UUCCUUGGCUGAAUUCGUAUGCGAGGCAGC ..........((.(((((((((...(.(((.....(((((......))))).(((((....-.)))))----......))).).....)))).))))).)). ( -28.10) >DroEre_CAF1 58064 96 - 1 AUUUAUGAAUCUC-CUCGAUCAUCGCCGCCACUUUUCUCUCUCUGCAGAAAUGCAGCUAAA-CGCUGC----UUCCUUGGCUGAAUUCGUAUGCGAGGCAGC ....((((.((..-...)))))).(((((((..((((((.....).))))).(((((....-.)))))----.....)))).....(((....))))))... ( -24.30) >DroYak_CAF1 57472 97 - 1 ACUUAUGAAUCUCCCUCGAUUAUCGCUGCCACUUUUCUCUCUCUGCAGAUAUGCAGCUAAA-CGCUGC----UUCCUUGGCUGAAUUCGUAUGCGAGGCAGC .......((((......))))...((((((...........((.((......(((((....-.)))))----.......)).))..(((....))))))))) ( -25.92) >DroAna_CAF1 52761 84 - 1 ------------------AUUCAGAGUACUACUUUUCUGAACCUGUACAAAUGCAGCUAAAAUGCCGCCGUGCUCCUUGGCUGAAUUCGUAUGCGAGGCAGC ------------------.(((((((........))))))).(((((....)))))......((((((((.......)))).....(((....))))))).. ( -22.60) >consensus AUUUAUGAAUCUCCCUCGAUACUUGCUGCCACUUUUCUCUCUCUGCAGAGAUGCAGCUAAA_CGCUGC____UUCCUUGGCUGAAUUCGUAUGCGAGGCAGC ........................((((((...........((.((.(((..(((((......)))))....)))....)).))..(((....))))))))) (-17.59 = -18.73 + 1.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:36:42 2006