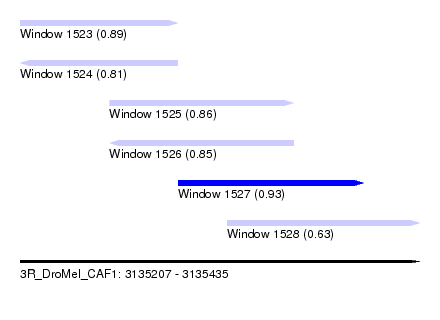
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,135,207 – 3,135,435 |
| Length | 228 |
| Max. P | 0.933932 |

| Location | 3,135,207 – 3,135,297 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -47.07 |
| Consensus MFE | -47.04 |
| Energy contribution | -46.85 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3135207 90 + 27905053 ACGCGCGCGAAAGUGGCCAACUGGUGGACAGUGGCCGAGUGGCCGGCCGUUUGGGCCAGGUUUAGUUUGGCUUAGCCUGGCUUGGACUUU ......((....))((((.((((.....))))((((....))))))))(((..(((((((((.((.....)).)))))))))..)))... ( -46.80) >DroSec_CAF1 10872 90 + 1 ACGCGCGCGAAAGUGGCCAACUGGUGGACAGUGGCCGAGUGGCCGGCCGUUUGGGCCAGGUUUAGUUUGGCUUAGCCUGGCUUGGACUUU ......((....))((((.((((.....))))((((....))))))))(((..(((((((((.((.....)).)))))))))..)))... ( -46.80) >DroSim_CAF1 11429 90 + 1 ACGCGCGCGAAAGUGGCCAACUGGUGGACAGUGGCCGAGUGGCCGGCCGUUUGGGCCAGGUUUAGUUUGGCUUAGCCUGGCUUGGACUUU ......((....))((((.((((.....))))((((....))))))))(((..(((((((((.((.....)).)))))))))..)))... ( -46.80) >DroEre_CAF1 11440 90 + 1 ACGCGCGCGAAAGUGGCCAACUGGUGGACAGUGGCCGAGUGGCCGGCCGUUUGGGCCAGGUUUAGCUUGGCUUAGCCUGGCUUGGACUUU ......((....))((((.((((.....))))((((....))))))))(((..(((((((((.(((...))).)))))))))..)))... ( -47.90) >consensus ACGCGCGCGAAAGUGGCCAACUGGUGGACAGUGGCCGAGUGGCCGGCCGUUUGGGCCAGGUUUAGUUUGGCUUAGCCUGGCUUGGACUUU ......((....))((((.((((.....))))((((....))))))))(((..(((((((((.(((...))).)))))))))..)))... (-47.04 = -46.85 + -0.19)

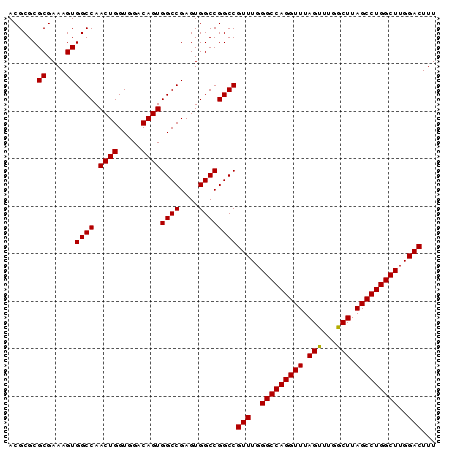
| Location | 3,135,207 – 3,135,297 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -30.12 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3135207 90 - 27905053 AAAGUCCAAGCCAGGCUAAGCCAAACUAAACCUGGCCCAAACGGCCGGCCACUCGGCCACUGUCCACCAGUUGGCCACUUUCGCGCGCGU (((((....((((((...((.....))...))))))......((((((((....))))((((.....)))).)))))))))......... ( -29.80) >DroSec_CAF1 10872 90 - 1 AAAGUCCAAGCCAGGCUAAGCCAAACUAAACCUGGCCCAAACGGCCGGCCACUCGGCCACUGUCCACCAGUUGGCCACUUUCGCGCGCGU (((((....((((((...((.....))...))))))......((((((((....))))((((.....)))).)))))))))......... ( -29.80) >DroSim_CAF1 11429 90 - 1 AAAGUCCAAGCCAGGCUAAGCCAAACUAAACCUGGCCCAAACGGCCGGCCACUCGGCCACUGUCCACCAGUUGGCCACUUUCGCGCGCGU (((((....((((((...((.....))...))))))......((((((((....))))((((.....)))).)))))))))......... ( -29.80) >DroEre_CAF1 11440 90 - 1 AAAGUCCAAGCCAGGCUAAGCCAAGCUAAACCUGGCCCAAACGGCCGGCCACUCGGCCACUGUCCACCAGUUGGCCACUUUCGCGCGCGU (((((....((((((...(((...)))...))))))......((((((((....))))((((.....)))).)))))))))......... ( -32.00) >consensus AAAGUCCAAGCCAGGCUAAGCCAAACUAAACCUGGCCCAAACGGCCGGCCACUCGGCCACUGUCCACCAGUUGGCCACUUUCGCGCGCGU (((((....((((((...((.....))...))))))......((((((((....))))((((.....)))).)))))))))......... (-30.12 = -30.12 + 0.00)

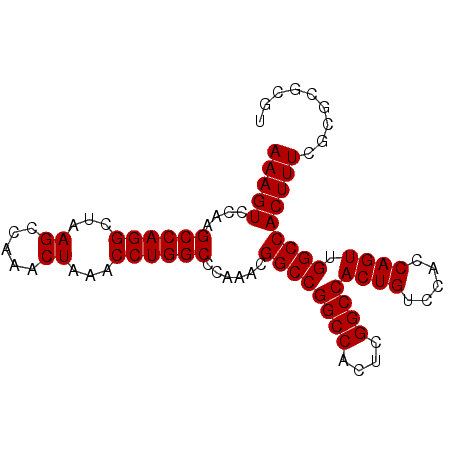
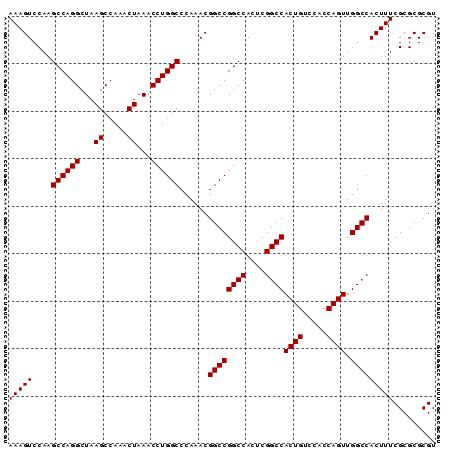
| Location | 3,135,258 – 3,135,363 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 98.57 |
| Mean single sequence MFE | -43.58 |
| Consensus MFE | -43.62 |
| Energy contribution | -43.25 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3135258 105 + 27905053 UGGGCCAGGUUUAGUUUGGCUUAGCCUGGCUUGGACUUUGCCUAUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCA .((((((((((.((.....)).))))))))))......((((....(((((.((...)).)))))))))(((((..(((((((.....)).)))))..))).)). ( -43.10) >DroSec_CAF1 10923 105 + 1 UGGGCCAGGUUUAGUUUGGCUUAGCCUGGCUUGGACUUUGCCUCUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCACAAAAAGUAGCUGCCAGCUCGCA .((((((((((.((.....)).))))))))))......((((....(((((.((...)).)))))))))(((((..(((((((.....)).)))))..))).)). ( -43.50) >DroSim_CAF1 11480 105 + 1 UGGGCCAGGUUUAGUUUGGCUUAGCCUGGCUUGGACUUUGCCUCUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCA (((((((((.((((...(((..((((......)).))..))).)))).)))))))))((.((.((((((((.((.....))((.....)).)))))).)))))). ( -43.50) >DroEre_CAF1 11491 105 + 1 UGGGCCAGGUUUAGCUUGGCUUAGCCUGGCUUGGACUUUGCCUCUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCA .((((((((((.(((...))).))))))))))......((((....(((((.((...)).)))))))))(((((..(((((((.....)).)))))..))).)). ( -44.20) >consensus UGGGCCAGGUUUAGUUUGGCUUAGCCUGGCUUGGACUUUGCCUCUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCA .((((((((((.(((...))).))))))))))......((((....(((((.((...)).)))))))))(((((..(((((((.....)).)))))..))).)). (-43.62 = -43.25 + -0.38)

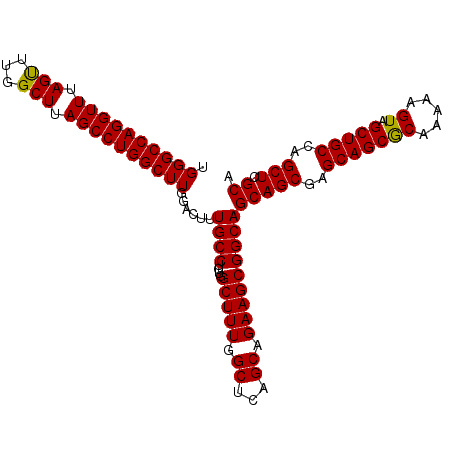
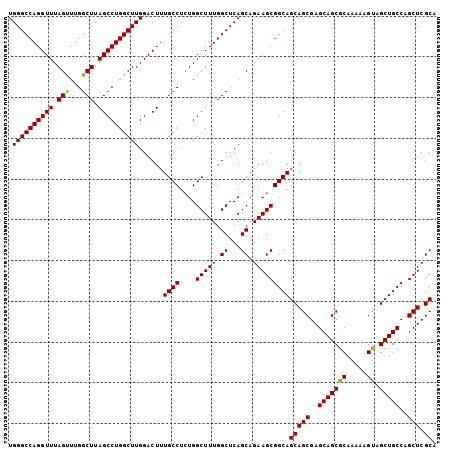
| Location | 3,135,258 – 3,135,363 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 98.57 |
| Mean single sequence MFE | -38.55 |
| Consensus MFE | -37.56 |
| Energy contribution | -37.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3135258 105 - 27905053 UGCGAGCUGGCAGCUACUUUUUGCGCUGCUCGCUGCUGCCGCUUCUGCUGAGCCAAAGCCAUAGGCAAAGUCCAAGCCAGGCUAAGCCAAACUAAACCUGGCCCA .((((((.(((.((........)))))))))))...((((((....))((.((....))))..))))........((((((...((.....))...))))))... ( -34.90) >DroSec_CAF1 10923 105 - 1 UGCGAGCUGGCAGCUACUUUUUGUGCUGCUCGCUGCUGCCGCUUCUGCUGAGCCAAAGCCAGAGGCAAAGUCCAAGCCAGGCUAAGCCAAACUAAACCUGGCCCA .((.(((.((((((.((.....)))))))).)))))....(((((((((.......)).))))))).........((((((...((.....))...))))))... ( -39.30) >DroSim_CAF1 11480 105 - 1 UGCGAGCUGGCAGCUACUUUUUGCGCUGCUCGCUGCUGCCGCUUCUGCUGAGCCAAAGCCAGAGGCAAAGUCCAAGCCAGGCUAAGCCAAACUAAACCUGGCCCA .((((((.(((.((........))))))))))).(((...(((((((((.......)).)))))))..)))....((((((...((.....))...))))))... ( -38.90) >DroEre_CAF1 11491 105 - 1 UGCGAGCUGGCAGCUACUUUUUGCGCUGCUCGCUGCUGCCGCUUCUGCUGAGCCAAAGCCAGAGGCAAAGUCCAAGCCAGGCUAAGCCAAGCUAAACCUGGCCCA .((((((.(((.((........))))))))))).(((...(((((((((.......)).)))))))..)))....((((((...(((...)))...))))))... ( -41.10) >consensus UGCGAGCUGGCAGCUACUUUUUGCGCUGCUCGCUGCUGCCGCUUCUGCUGAGCCAAAGCCAGAGGCAAAGUCCAAGCCAGGCUAAGCCAAACUAAACCUGGCCCA .((((((.(((.((........))))))))))).(((...(((((((((.......)).)))))))..)))....((((((...((.....))...))))))... (-37.56 = -37.62 + 0.06)

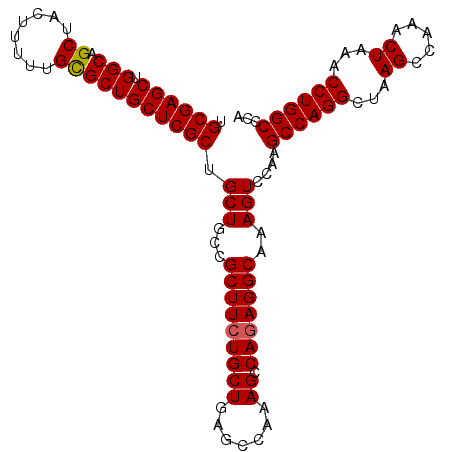
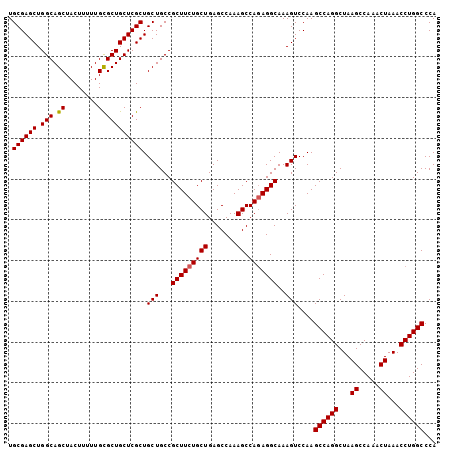
| Location | 3,135,297 – 3,135,403 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 99.06 |
| Mean single sequence MFE | -39.52 |
| Consensus MFE | -38.96 |
| Energy contribution | -38.77 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
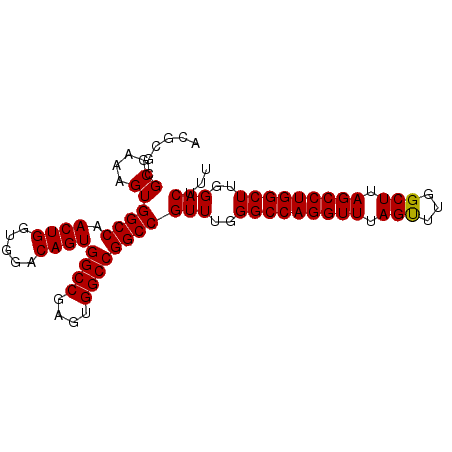
>3R_DroMel_CAF1 3135297 106 + 27905053 GCCUAUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAA .....(((((...(((((((((((((((((((.((.....))((.....)).)))))).))((....)).)))((....))..))))))))..)))))........ ( -39.00) >DroSec_CAF1 10962 106 + 1 GCCUCUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCACAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAA ...(((((((...(((((((((((((((((((.((.....))((.....)).)))))).))((....)).)))((....))..))))))))..))))).))..... ( -39.50) >DroSim_CAF1 11519 106 + 1 GCCUCUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAA ...(((((((...(((((((((((((((((((.((.....))((.....)).)))))).))((....)).)))((....))..))))))))..))))).))..... ( -39.80) >DroEre_CAF1 11530 106 + 1 GCCUCUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAA ...(((((((...(((((((((((((((((((.((.....))((.....)).)))))).))((....)).)))((....))..))))))))..))))).))..... ( -39.80) >consensus GCCUCUGGCUUUGGCUCAGCAGAAGCGGCAGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAA .....(((((...(((((((((((((((((((.((.....))((.....)).)))))).))((....)).)))((....))..))))))))..)))))........ (-38.96 = -38.77 + -0.19)

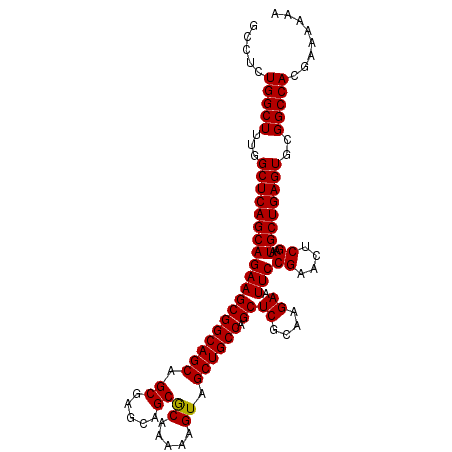
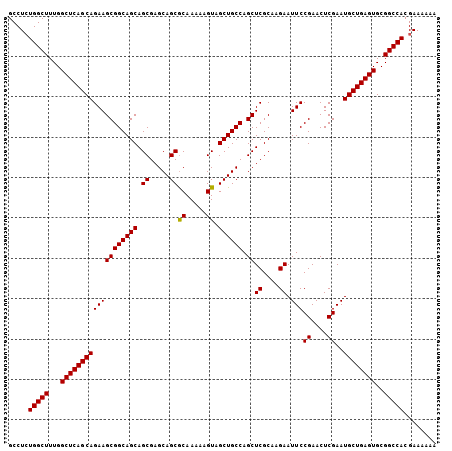
| Location | 3,135,325 – 3,135,435 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -26.85 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3135325 110 + 27905053 C--AGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAAACAUUCAAAUGCCUCGCA--------CUCGCCGCCAAAUC (--((((((((((.((((........)).))..))))))........((....))..))))).(.(((((...(((.......)))...(((...)))--------...))))))..... ( -32.10) >DroPse_CAF1 9119 119 + 1 CACAGCAGC-AGCAGGCAAAAAAGUUGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAAACAUUCAAAUGCCUCGCACGGUACGGCUCGCCGCCAAAUC ....(((((-(((..........))))))))((((((....))....((....))...)))).(.((((((..(((.......)))...((((......)))).))).))))........ ( -34.10) >DroSec_CAF1 10990 110 + 1 C--AGCAGCGAGCAGCACAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAAACAUUCAAAAGCCUCGCA--------CUCGCCGCCAAAUC .--.((.(((((((((((.....)).)))))((((((....))....((....))...)))).(((((((...(((.......)))....)))..)))--------))))).))...... ( -32.30) >DroSim_CAF1 11547 110 + 1 C--AGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAAACAUUCAAAUGCCUCGCA--------CUCGCCGCCAAAUC (--((((((((((.((((........)).))..))))))........((....))..))))).(.(((((...(((.......)))...(((...)))--------...))))))..... ( -32.10) >DroPer_CAF1 9092 119 + 1 CACAGCAGC-AGCAGGCAAAAAAGUUGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAAACAUUCAAAUGCCUCGCACGGUACGGCUCGCCGCCAAAUC ....(((((-(((..........))))))))((((((....))....((....))...)))).(.((((((..(((.......)))...((((......)))).))).))))........ ( -34.10) >DroEre_CAF1 11558 110 + 1 C--AGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAAACAUUCAAAUGCCUCGCA--------CUCGCCGCCAAAUC (--((((((((((.((((........)).))..))))))........((....))..))))).(.(((((...(((.......)))...(((...)))--------...))))))..... ( -32.10) >consensus C__AGCAGCGAGCAGCGCAAAAAGUAGCUGCCAGCUCGCAAGAAUUCCGAACUCGAAUGCUGAGUGCGGCCACGAAAAAAACAUUCAAAUGCCUCGCA________CUCGCCGCCAAAUC ....((.(((((((((..........)))))((((((....))....((....))...)))).(((.(((...(((.......)))....))).)))..........)))).))...... (-26.85 = -27.52 + 0.67)

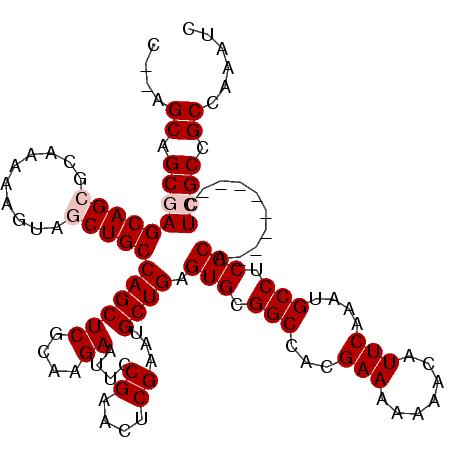
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:31 2006