
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,179,557 – 25,179,698 |
| Length | 141 |
| Max. P | 0.933732 |

| Location | 25,179,557 – 25,179,666 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.19 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -12.81 |
| Energy contribution | -16.23 |
| Covariance contribution | 3.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
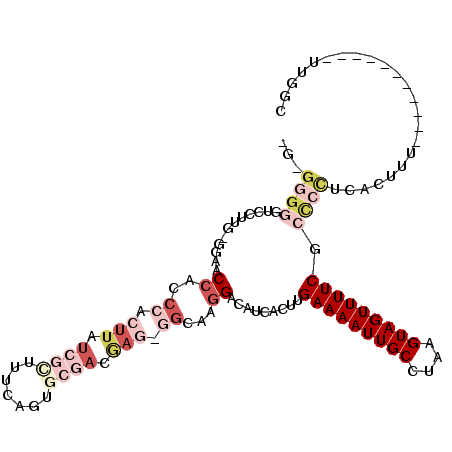
>3R_DroMel_CAF1 25179557 109 + 27905053 -GUGGGGGUCCUUG-GGAACCACCCACUUAUCGUUUUCAGUGCGACGAGCGGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCCCCUCACUUUU-----UUUUUUUGGC -(((((((((((((-(....)..((.((..((((.......))))..)).))))))))).......(((((((((....)))))))))...))))))....-----.......... ( -36.20) >DroSim_CAF1 38234 103 + 1 -G-GGGGGUCCUUG-GGCACCACCCACUUAUCGCUUUCAGUGCGACGAGCGGCAAGGACAUCACUUGAAAAUUGCGUAAGUAGUUUUCGCCCCUCACUUU----------UUUGGC -(-(((((((((((-(....)..((.((..((((.......))))..)).))))))))).......(((((((((....)))))))))..))))).....----------...... ( -41.50) >DroEre_CAF1 38739 102 + 1 ---GGGGGUGCUUG-GGAACCACCCACUUAUCGCUUUCAGUGCGACGAGAGGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCGCCCCUUACUUUCU----------UGGC ---(((((((((((-((.....))).((..((((.......))))..)))))).............(((((((((....)))))))))))))))........----------.... ( -33.60) >DroYak_CAF1 40096 115 + 1 CGAGGGGAUCCUUG-GGAACCACCCACUUAUCGCUUUCAGUGCGAGGAGAGGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCGCCCCUCACUUUCUUUCUUUUUUUUGGC .((((((.((((((-((......)).(((.((((.......)))).)))...))))))........(((((((((....))))))))).))))))..................... ( -37.50) >DroAna_CAF1 36204 88 + 1 -A-------------GGACCCCUCCACUUAUCGCUUUUAGUUGAACAA--CGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCUUCUCAGUU------------UUAGC -.-------------(((....))).......(((...(..(((....--(....)..........(((((((((....))))))))).....)))..)------------..))) ( -14.70) >DroPer_CAF1 42827 85 + 1 -GUGGG-----UGGGGUAGCAGCCCACUUAUCGCGUCCAGC-------------AGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCU-UUCACUUUU-----------UGGU -(((((-----((((((....)))).........((((...-------------.))))..)))).(((((((((....)))))))))...-..)))....-----------.... ( -24.20) >consensus _G_GGGGGUCCUUG_GGAACCACCCACUUAUCGCUUUCAGUGCGACGAG_GGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCGCCCCUCACUUU___________UUGGC ...((((............((..((.(((.((((.......)))).))).))...)).........(((((((((....))))))))).))))....................... (-12.81 = -16.23 + 3.42)

| Location | 25,179,595 – 25,179,698 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.72 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -11.52 |
| Energy contribution | -11.92 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25179595 103 + 27905053 UGCGACGAGCGGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCCCCUCACUUUU--------UUUUUUUGGCCUUGU-CCGCAUUGUUUUAUUCCGUUUCCCUUC- ...((((((((((((((.........(((((((((....)))))))))...((........--------.......))))))).-)))).))))).................- ( -24.46) >DroSim_CAF1 38271 98 + 1 UGCGACGAGCGGCAAGGACAUCACUUGAAAAUUGCGUAAGUAGUUUUCGCCCCUCACUUU-------------UUUGGCCUUGU-CCGCAUUGUUUUAUUCCGUUUCCCUUU- ...((((((((((((((.........(((((((((....)))))))))...((.......-------------...))))))).-)))).))))).................- ( -28.60) >DroEre_CAF1 38775 98 + 1 UGCGACGAGAGGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCGCCCCUUACUUUC---U----------UGGCCUUGU-CCGCAUUGUUUUAUUCUGUUUCCCUUU- (((((((((.(.((((((.......((((((((((....)))))))))).........)))---)----------)).))))))-.))))......................- ( -25.09) >DroWil_CAF1 117432 102 + 1 CA--------CACAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCUUC-AACCAUUUUCCUUCAUGUUUUUGUUGUGUU-UCUUCAUGUUUUGUUGUUAUUUUCGUU- ((--------(((((((((((.....(((((((((....))))))))).....-..............))))))))).))))..-...........................- ( -15.75) >DroYak_CAF1 40135 108 + 1 UGCGAGGAGAGGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCGCCCCUCACUUUC---UUUCUUUUUUUUGGCCUUGU-CCGCAUUGUUUUAUUCUGUUUCCCUUU- ...((((.((((((.(((((......(((((((((....)))))))))(((..........---............)))..)))-))(.((......)).))))))))))).- ( -23.45) >DroAna_CAF1 36230 96 + 1 UUGAACAA--CGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCUCUUCUCAGUU---------------UUAGCCUUGUUUUGCUUUGUUUUAUAUUGUUUCCUUUCU ..((((((--.(((((.(((......(((((((((....)))))))))........((.---------------...))..)))))))).))))))................. ( -16.30) >consensus UGCGACGAG_GGCAAGGACAUCACUUGAAAAUUGCCUAAGUAGUUUUCGCCCCUCACUUU_____________UUUGGCCUUGU_CCGCAUUGUUUUAUUCUGUUUCCCUUU_ .(((......((((((.......)))(((((((((....))))))))).............................)))......)))........................ (-11.52 = -11.92 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:36:32 2006