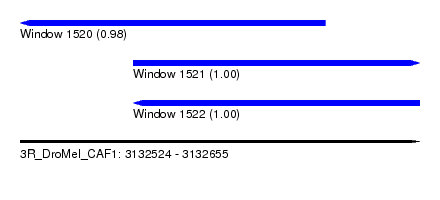
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,132,524 – 3,132,655 |
| Length | 131 |
| Max. P | 0.999156 |

| Location | 3,132,524 – 3,132,624 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -20.07 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
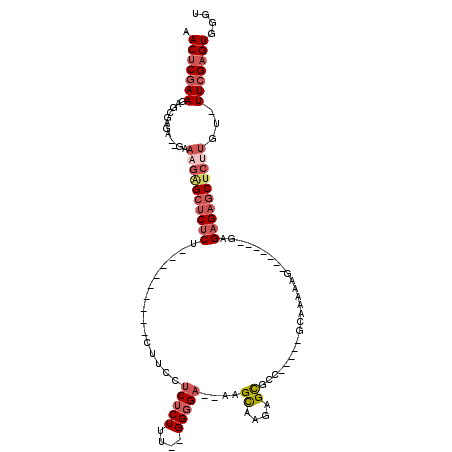
>3R_DroMel_CAF1 3132524 100 - 27905053 CUUUCUCUCUUC--GGGGA--AAGCAAGAGCACC-----GCAAAACG-------GAGAGAUCUCUUGU-UUCGAGUGGGUUUUA---AUGGUAAAACGCUCAAGCUUUUGCGUCGCUUUU ((((((((....--)))))--))).((((((..(-----((((((.(-------(((....)))).((-((.(((((..((...---.....))..))))))))))))))))..)))))) ( -32.70) >DroSec_CAF1 7925 100 - 1 CUUCCUCUCUUU--GGGGA--AAGCAAGAGCGCC-----GCAAAAAG-------GAGAGAGCUCUUGU-UUCGAGUGGGUUUUU---GGAGGAAAACGCUCAAGCUUUUGCGUCGCUUUU .((((((.....--)))))--)...(((((((.(-----((((((((-------(((....)))))((-((.(((((..((((.---...))))..)))))))))))))))).))))))) ( -37.60) >DroSim_CAF1 8549 100 - 1 CUUCCUCUCUUU--GGGGA--AAGCAAGAGCGCC-----GCAAAAAG-------GAGAGAGCUCUAGU-UUCGAGUGGGUUUUU---AGGGGAAAACGCUCAAGCUUUUGCGUCGCUUUU .((((((.....--)))))--)...(((((((.(-----((((((.(-------(((....)))).((-((.(((((..((((.---...))))..)))))))))))))))).))))))) ( -37.20) >DroPer_CAF1 6702 120 - 1 CCCGCUCUCUUAUGGGGGAGUGUGUAAGAGUGCCUGAGCGCCAGAGGGCAGUGGAAGAGAGCGAGUGCUUUCGAGUGGCAUUUUUCGGGGGAAAAACACUCAAGCUUUGGCGGGGGGAUU ((((((((((....)))))))..........(((.(((((((....))).......((((((....))))))(((((...((((((....)))))))))))..)))).))))))...... ( -45.30) >DroEre_CAF1 8323 100 - 1 CCUUUUCUCUUU--GGCGC--AUGCAAGAGCGCG-----CCCAA-AA-------GAGAGAGCUCUUGUUUUGGAGUGGUUUUUU---GGGGGAAAACGCUCAAGCUUUUGCGUCGCUCUU ............--(((((--(.((..(((((..-----(((..-((-------((((..(((((......)))))..))))))---..)))....)))))..))...))))))...... ( -36.90) >DroYak_CAF1 8335 101 - 1 CGUCUUCUCUUU--GGGGA--AUGCAAGAGCGCC-----CCCAAAAA-------GAGAGAGCUCAUGGGUUCGAGUGGUUUUUU---GGGGGAAAACGCUCAAGCUUUUGCGUCGCUCUU .((.(((((...--.))))--).))(((((((((-----((((((((-------(...(((((....)))))......))))))---)))))...((((..........))))))))))) ( -40.40) >consensus CUUCCUCUCUUU__GGGGA__AAGCAAGAGCGCC_____GCAAAAAG_______GAGAGAGCUCUUGU_UUCGAGUGGGUUUUU___GGGGGAAAACGCUCAAGCUUUUGCGUCGCUUUU .....(((((....)))))....((((((((.......................(((....)))........(((((..(((((.....)))))..)))))..))))))))......... (-20.07 = -20.77 + 0.70)
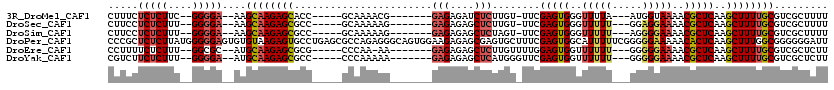
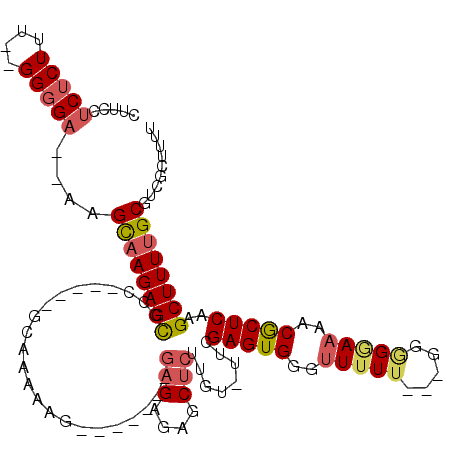
| Location | 3,132,561 – 3,132,655 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.17 |
| Mean single sequence MFE | -36.71 |
| Consensus MFE | -11.98 |
| Energy contribution | -13.60 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.24 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.33 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3132561 94 + 27905053 ACCCACUCGAA-ACAAGAGAUCUCUC-------CGUUUUGC-----GGUGCUCUUGCUU--UCCCC--GAAGAGAGAAAG---------AGAGAGCUCUUUCACUGACGCUCUUCGAGUU ....(((((((-(((((((......(-------((.....)-----))..)))))).))--))...--((((((.(((((---------((....))))))).......)))))))))). ( -31.30) >DroSec_CAF1 7962 92 + 1 ACCCACUCGAA-ACAAGAGCUCUCUC-------CUUUUUGC-----GGCGCUCUUGCUU--UCCCC--AAAGAGAGGAAG---------AGAGAGCUCUUUC--UCACGCUCUUCGAGUU ....(((((((-..((((((((((((-------(((((((.-----((.((....))..--.)).)--)))))).....)---------)))))))))))..--........))))))). ( -33.40) >DroSim_CAF1 8586 92 + 1 ACCCACUCGAA-ACUAGAGCUCUCUC-------CUUUUUGC-----GGCGCUCUUGCUU--UCCCC--AAAGAGAGGAAG---------AGAGAGCUCUUUC--UCGCGCUCUUCGAGUU ....(((((((-...(((((((((((-------(((((((.-----((.((....))..--.)).)--)))))).....)---------))))))))))...--........))))))). ( -33.04) >DroPer_CAF1 6742 118 + 1 UGCCACUCGAAAGCACUCGCUCUCUUCCACUGCCCUCUGGCGCUCAGGCACUCUUACACACUCCCCCAUAAGAGAGCGGGGGGUGUGUGUGUGUGCUCUUUC--AUUUGCUCUUCGUGUU ....((.((((((((................(((....))).....(((((.(.(((((((((((((........).)))))))))))).).))))).....--...)))).)))).)). ( -41.60) >DroEre_CAF1 8360 92 + 1 AACCACUCCAAAACAAGAGCUCUCUC-------UU-UUGGG-----CGCGCUCUUGCAU--GCGCC--AAAGAGAAAAGG---------AGAGAGCCCUUUC--UCUCGCUCUUCGAGUU ....((((......((((((..((((-------((-...((-----(((((....))..--)))))--.))))))....(---------((((((....)))--)))))))))).)))). ( -34.80) >DroYak_CAF1 8372 97 + 1 AACCACUCGAACCCAUGAGCUCUCUC-------UUUUUGGG-----GGCGCUCUUGCAU--UCCCC--AAAGAGAAGACG-----AGAGAGAGAGCCCUUUC--UCUCGCUCUUCGAGUU ....(((((((.....(((((((.((-------((((((((-----((.((....))..--)))))--)))))))))).(-----((((((((....)))))--))))))))))))))). ( -46.10) >consensus ACCCACUCGAA_ACAAGAGCUCUCUC_______CUUUUGGC_____GGCGCUCUUGCAU__UCCCC__AAAGAGAGGAAG_________AGAGAGCUCUUUC__UCUCGCUCUUCGAGUU ....(((((((...(((.(((((((........(((((.((......)).(((((..............))))).))))).........))))))).)))............))))))). (-11.98 = -13.60 + 1.62)
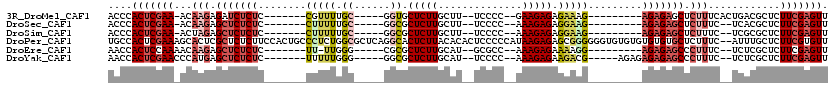
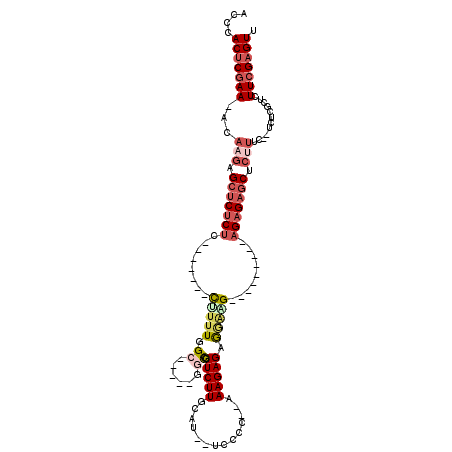
| Location | 3,132,561 – 3,132,655 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.17 |
| Mean single sequence MFE | -38.57 |
| Consensus MFE | -17.30 |
| Energy contribution | -18.86 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3132561 94 - 27905053 AACUCGAAGAGCGUCAGUGAAAGAGCUCUCU---------CUUUCUCUCUUC--GGGGA--AAGCAAGAGCACC-----GCAAAACG-------GAGAGAUCUCUUGU-UUCGAGUGGGU .((((((((((.(((.........(((((..---------((((((((....--)))))--)))..))))).((-----(.....))-------)...)))))))...-.)))))).... ( -32.50) >DroSec_CAF1 7962 92 - 1 AACUCGAAGAGCGUGA--GAAAGAGCUCUCU---------CUUCCUCUCUUU--GGGGA--AAGCAAGAGCGCC-----GCAAAAAG-------GAGAGAGCUCUUGU-UUCGAGUGGGU .((((((((..(....--).(((((((((((---------(((((((.....--)))))--..((....))...-----.......)-------)))))))))))).)-))))))).... ( -40.00) >DroSim_CAF1 8586 92 - 1 AACUCGAAGAGCGCGA--GAAAGAGCUCUCU---------CUUCCUCUCUUU--GGGGA--AAGCAAGAGCGCC-----GCAAAAAG-------GAGAGAGCUCUAGU-UUCGAGUGGGU .((((((((..(....--)..((((((((((---------(((((((.....--)))))--..((....))...-----.......)-------)))))))))))..)-))))))).... ( -38.60) >DroPer_CAF1 6742 118 - 1 AACACGAAGAGCAAAU--GAAAGAGCACACACACACACCCCCCGCUCUCUUAUGGGGGAGUGUGUAAGAGUGCCUGAGCGCCAGAGGGCAGUGGAAGAGAGCGAGUGCUUUCGAGUGGCA ..(((.....((....--(..((.((((.(..((((((((((((........)))))).))))))..).))))))...)(((....))).))....((((((....))))))..)))... ( -42.60) >DroEre_CAF1 8360 92 - 1 AACUCGAAGAGCGAGA--GAAAGGGCUCUCU---------CCUUUUCUCUUU--GGCGC--AUGCAAGAGCGCG-----CCCAA-AA-------GAGAGAGCUCUUGUUUUGGAGUGGUU .((((.((((((((((--((......)))))---------)..(((((((((--(((((--..((....)))))-----))..)-))-------))))))))))).....).)))).... ( -37.00) >DroYak_CAF1 8372 97 - 1 AACUCGAAGAGCGAGA--GAAAGGGCUCUCUCUCU-----CGUCUUCUCUUU--GGGGA--AUGCAAGAGCGCC-----CCCAAAAA-------GAGAGAGCUCAUGGGUUCGAGUGGUU .((((((((((.((((--((......)))))).))-----)(..((((((((--((((.--.(((....))).)-----))))..))-------)))))..).......))))))).... ( -40.70) >consensus AACUCGAAGAGCGAGA__GAAAGAGCUCUCU_________CUUCCUCUCUUU__GGGGA__AAGCAAGAGCGCC_____GCAAAAAG_______GAGAGAGCUCUUGU_UUCGAGUGGGU .(((((((............((((((((((...............(((((....)))))....((....)).........................))))))))))...))))))).... (-17.30 = -18.86 + 1.56)
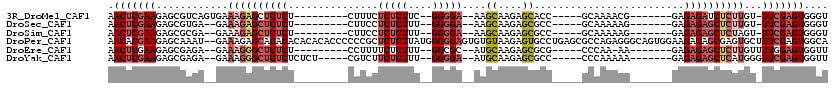
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:26 2006