
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,148,638 – 25,148,770 |
| Length | 132 |
| Max. P | 0.670138 |

| Location | 25,148,638 – 25,148,746 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -23.33 |
| Energy contribution | -23.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25148638 108 + 27905053 UUUUUUUUUCA-------GUGCGCGUGUUUGUUUGUUUAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCC-AGG .....((((((-------((..(((....)))..((((.((((.((((((((((.....))))))))))...))))..)))))))))))).......((((.((....))))-.)) ( -29.30) >DroVir_CAF1 7009 106 + 1 GUG-UGUGUGU-------GC-CUGGUGUUUGUUUGUUUAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCA-CAG .((-((((..(-------(.-((((((...((((.....((((.((((((((((.....))))))))))...))))(....).....))))....)))))).))....))))-)). ( -33.00) >DroPse_CAF1 7737 111 + 1 ----CCCUGCAUACGUAUAUGUAUAUGUUUGUUUGCUUAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCC-AGG ----.((((((.((((((....))))))(((((((....((((.((((((((((.....))))))))))...))))(....)................)))))))...)).)-))) ( -30.50) >DroGri_CAF1 87567 95 + 1 -------------------U-UUGGUGUUUGUUUGUUUAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUACC-CAG -------------------.-..((((.(((((((....((((.((((((((((.....))))))))))...))))(....)................)))))))...))))-... ( -27.70) >DroAna_CAF1 6471 107 + 1 UU--UUUUGUA-------GUGUUGGUGUUUGUUUGUUUAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCCGAGG ..--.......-------...((((((.(((((((....((((.((((((((((.....))))))))))...))))(....)................)))))))...)))))).. ( -29.10) >DroPer_CAF1 8747 111 + 1 ----CCCUGCAUAUGUAUAUGUAUAUGUUUGUUUGCUUAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCC-AGG ----.((((((((((((....)))))))(((((((....((((.((((((((((.....))))))))))...))))(....)................)))))))....).)-))) ( -29.80) >consensus ____UCUUGCA_______GUGUUGGUGUUUGUUUGUUUAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCC_AGG ............................(((((((....((((.((((((((((.....))))))))))...))))(....)................)))))))........... (-23.33 = -23.33 + -0.00)

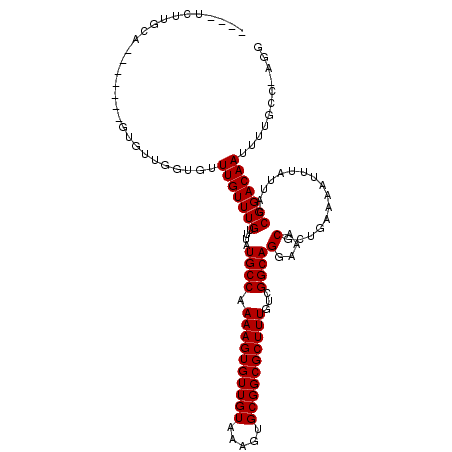

| Location | 25,148,668 – 25,148,770 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25148668 102 + 27905053 UAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCCAGGGGCAUAAAAAA----------GAGAGAUCCCCAC---- ((((((.((((((((((.....)))))))))).(((((.(....))))))..........((((.((....)))).)))))))).....----------.............---- ( -29.90) >DroVir_CAF1 7037 110 + 1 UAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCACAGGCCAAAAAAAAAAAAAAAAAGCCGAG-AGACAA----- ..((((.((((((((((.....)))))))))).(((((.(....))))))............)).))..........(((..................)))...-......----- ( -23.77) >DroGri_CAF1 87584 100 + 1 UAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUACCCAG-GCAUAAAAAA----------GAGUA-AGAGAAU---- ((((((.((((((((((.....)))))))))).(((((.(....))))))...........................)-))))).....----------.....-.......---- ( -27.10) >DroWil_CAF1 81561 99 + 1 UAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAGCGCUGAAAAUUUAUUACCGGACAAUUUUGCCAUG-GCAUAAAAAA----------AAGCG-CCG-ACC---- (((((((((((((((((.....))))))))))...(((((((....(((............)))....))))))).))-))))).....----------.....-...-...---- ( -29.30) >DroMoj_CAF1 6493 99 + 1 UAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGC-CAGGGCAUAAAAAA----------GCGAG-AGACAA----- ((((((.((((((((((.....))))))))))...(((((((....(((............)))....))))))-)..)))))).....----------((...-.).)..----- ( -28.40) >DroPer_CAF1 8780 101 + 1 UAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCCAGGGG-AUAAAAAA----------GAG---CCG-ACUGUGG ....(((((((((((((.....))))))))))((((((.(....)........(((((..((((.((....)))).))..-)))))...----------..)---)))-))..))) ( -32.80) >consensus UAUGCCAAAAGUGUUGUAAAGUGCGGCGCUUUGUCGGCAGGAAACGCUGAAAAUUUAUUACCGGACAAUUUUGCCAAGGGCAUAAAAAA__________GAGAG_ACACAA_____ ((((((.((((((((((.....)))))))))).(((((.(....))))))............................))))))................................ (-21.38 = -22.22 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:36:17 2006