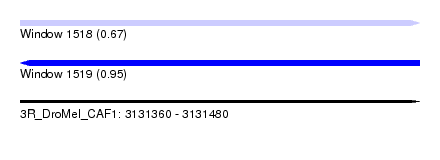
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,131,360 – 3,131,480 |
| Length | 120 |
| Max. P | 0.946273 |

| Location | 3,131,360 – 3,131,480 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -11.23 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3131360 120 + 27905053 UCCAUUGAAUAAUAUUCAUCUUGUUUGAUCCAAGUAGAUCAAAGAGUAAAUUAAAAGUUACUUAUAGAUCGAUGUGGAAAAGUAGGAGAUCAAAUCUUAUUAAGAACUUUUUUAGUUCUA ....(..(((((........)))))..)((((....((((...((((((........))))))...))))....))))..(((((((.......))))))).((((((.....)))))). ( -25.00) >DroSec_CAF1 6802 120 + 1 ACCAUUGAAUAAUAUUCCCCUUGUUUGAUCCAAGUAGAUCAAAGAGUUAAUUGAAAGAUACUUAUAGAUCGAAGUUGAAAAGUGGGGGAUCGAAUCUUAUUAAGAACUAUUUUAGUUCUA .......(((((.((((((((..(((....(((.(.((((...((((..(((....)))))))...))))..).)))..)))..))))...)))).))))).(((((((...))))))). ( -25.90) >DroSim_CAF1 7365 120 + 1 ACCAUUGAAUAAUAUUCCCCUUGUUUGAUCCAAGUAGAUCAAAGAGUAAAUUGAAAGAUACUUAUAGAUCGAAGUUGAAAAGUGGGGGAUCGAAUCUUAUUAAGAACUAUUUUAGUUCUA .......(((((.((((((((..(((....(((.(.((((...(((((..((....)))))))...))))..).)))..)))..))))...)))).))))).(((((((...))))))). ( -26.10) >DroEre_CAF1 7059 116 + 1 ACUGUUGAAUAAUAUUUCCCUUGUUUGAUCUAAGCAGAUCGAUGGG-AAACUAAAAGAUACUUAUAGAUCGAAGCUAAGAAGUGAGAGCUCGAAUC---UUAAGAUCUUUUUUAGUUGUA ........(((((.((((((....(((((((....))))))).)))-)))..(((((((.((((.(((((((.(((..........)))))).)))---))))))))))))...))))). ( -30.00) >DroYak_CAF1 7143 91 + 1 -------------GUUUCAUCUGUUUGAUCUGAGUAGAUCAAUGGGGAAAUUAAAA----------GAUCGAA------AAGUGAGAGAUCGAAUCGUAUUAAGAACUAUUUUAGUUCUA -------------(((((.((((((.(((((....)))))))))))))))).....----------((((...------........))))...........(((((((...))))))). ( -18.70) >consensus ACCAUUGAAUAAUAUUCCCCUUGUUUGAUCCAAGUAGAUCAAAGAGUAAAUUAAAAGAUACUUAUAGAUCGAAGUUGAAAAGUGGGAGAUCGAAUCUUAUUAAGAACUAUUUUAGUUCUA ....................((.(((((((......))))))).))....................((((.................))))...........((((((.....)))))). (-11.23 = -11.67 + 0.44)

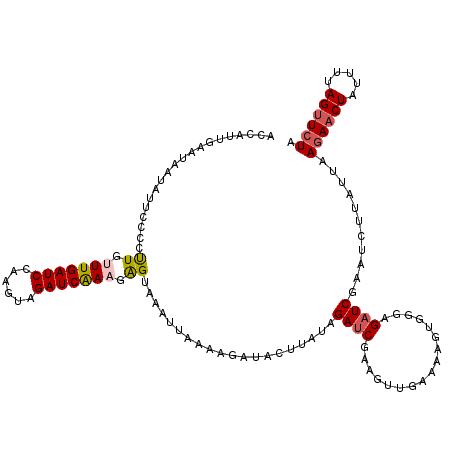
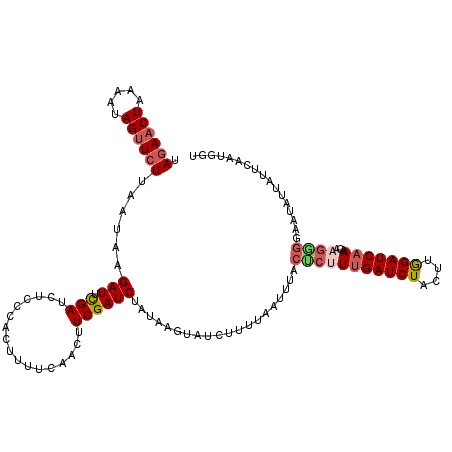
| Location | 3,131,360 – 3,131,480 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -9.77 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3131360 120 - 27905053 UAGAACUAAAAAAGUUCUUAAUAAGAUUUGAUCUCCUACUUUUCCACAUCGAUCUAUAAGUAACUUUUAAUUUACUCUUUGAUCUACUUGGAUCAAACAAGAUGAAUAUUAUUCAAUGGA .((((((.....)))))).(((((...((.((((........((((....((((.(..(((((........)))))..).))))....)))).......)))).))..)))))....... ( -18.76) >DroSec_CAF1 6802 120 - 1 UAGAACUAAAAUAGUUCUUAAUAAGAUUCGAUCCCCCACUUUUCAACUUCGAUCUAUAAGUAUCUUUCAAUUAACUCUUUGAUCUACUUGGAUCAAACAAGGGGAAUAUUAUUCAAUGGU .(((((((...))))))).............(((((.((((.((......)).....))))................((((((((....))))))))...)))))............... ( -21.00) >DroSim_CAF1 7365 120 - 1 UAGAACUAAAAUAGUUCUUAAUAAGAUUCGAUCCCCCACUUUUCAACUUCGAUCUAUAAGUAUCUUUCAAUUUACUCUUUGAUCUACUUGGAUCAAACAAGGGGAAUAUUAUUCAAUGGU .(((((((...))))))).............(((((.((((.((......)).....))))................((((((((....))))))))...)))))............... ( -21.00) >DroEre_CAF1 7059 116 - 1 UACAACUAAAAAAGAUCUUAA---GAUUCGAGCUCUCACUUCUUAGCUUCGAUCUAUAAGUAUCUUUUAGUUU-CCCAUCGAUCUGCUUAGAUCAAACAAGGGAAAUAUUAUUCAACAGU .........((((((((((((---((((.(((((..........))))).))))).)))).))))))).((((-(((...(((((....)))))......)))))))............. ( -27.80) >DroYak_CAF1 7143 91 - 1 UAGAACUAAAAUAGUUCUUAAUACGAUUCGAUCUCUCACUU------UUCGAUC----------UUUUAAUUUCCCCAUUGAUCUACUCAGAUCAAACAGAUGAAAC------------- .(((((((...)))))))........(((((((........------...))))----------..............(((((((....)))))))......)))..------------- ( -14.10) >consensus UAGAACUAAAAUAGUUCUUAAUAAGAUUCGAUCUCCCACUUUUCAACUUCGAUCUAUAAGUAUCUUUUAAUUUACUCUUUGAUCUACUUGGAUCAAACAAGGGGAAUAUUAUUCAAUGGU .((((((.....))))))......(((.(((.................))))))....................(((((((((((....)))))))...))))................. ( -9.77 = -10.73 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:23 2006