| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,121,978 – 25,122,098 |
| Length | 120 |
| Max. P | 0.619614 |

| Location | 25,121,978 – 25,122,098 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -30.22 |
| Energy contribution | -31.50 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25121978 120 + 27905053 UGGCUCGACAGACUUGUGGGGCACGGCAGCCAGUUUCUUCCGUAUGCUCAGUAGGAGCUUUGCCGAACUGGAAAGACCCUUUUCCAGGGCCCUCACCUCGUCUGAACUGGGUUCGAAUUU .((((((.(((((..(((((((.((((((..(((((((.(.(......).).)))))))))))))..((((((((....)))))))).)))).)))...)))))...))))))....... ( -45.10) >DroSec_CAF1 50485 120 + 1 UGGAUCAGCAGACUUGUAGGGCACGGCAUCCACUUUCUUUCGGAUGCUUAGUAGAAGCUUUGCCGAACUGGAAAGACCCUUCUCCAGUGCCCUCACCUCGUCUGAACUGGGUUCGAAUUC .(((((..(((((..((((((((((((((((..........)))))))..(.(((((((((.((.....))))))...))))).).))))))).))...))))).....)))))...... ( -41.50) >DroSim_CAF1 53032 120 + 1 UGGCUCGGCAGACAUGUAGGGUACGGCAUCCACUUUCUUUCGGAUGCUUAGUAGAAGCUUUGCCGAACUGGAAAGACCCUUCUCCAGUGCCCUCACCUCGUCUGAACUGGGUUCGAAUUC .((((((((((((..((((((((((((((((..........)))))))..(.(((((((((.((.....))))))...))))).).))))))).))...)))))..)))))))....... ( -40.70) >DroEre_CAF1 53162 120 + 1 UGGCCUGGCGGAUUUGCAGGCCACUGCAGUCAGUUUCUGCCGCAUGCUCAGCAGGAGCUUUGCCGCCCUGGAAAGACCCUUUUCCAGUGCCCGCAUCUCGUCCAAACUGGGACUGGAUUU .(((..(((((..((((((....))))))..(((((((((.(......).)))))))))...)))))((((((((....)))))))).)))...((((.((((......)))).)))).. ( -47.60) >DroYak_CAF1 52484 120 + 1 UGGCAUGGCAGAUAUGCAGGGCACUGCAGUCAGUUUCUUUCGCAUGCUCAGUAGGAGCUUUGCCGACCUGGAAAGUCCCUUUUCCAGUGCCCUCAUCUCGUCUAAACUGGGUAUGGAUUU .(((((((((((.((((((((.((((....)))).))))..))))((((.....)))))))))))..((((((((....)))))))))))).((((((((.......)))).)))).... ( -37.00) >consensus UGGCUCGGCAGACUUGUAGGGCACGGCAGCCAGUUUCUUUCGCAUGCUCAGUAGGAGCUUUGCCGAACUGGAAAGACCCUUUUCCAGUGCCCUCACCUCGUCUGAACUGGGUUCGAAUUU .((((((((((((.((.(((((.(((((...(((((((..............))))))).)))))..((((((((....)))))))).)))))))....)))))..)))))))....... (-30.22 = -31.50 + 1.28)

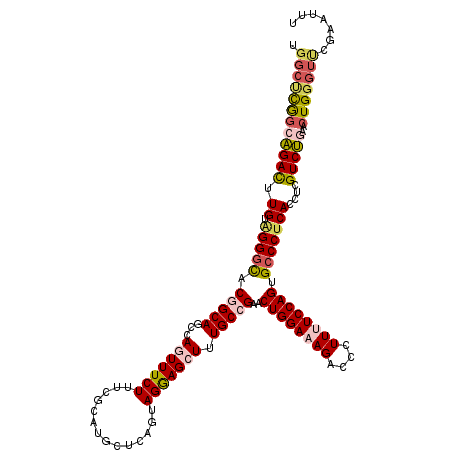
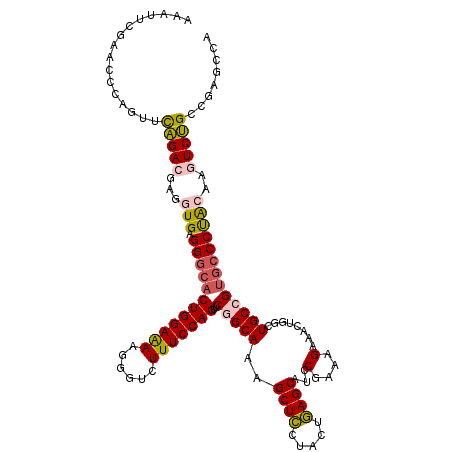
| Location | 25,121,978 – 25,122,098 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -41.42 |
| Consensus MFE | -29.84 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25121978 120 - 27905053 AAAUUCGAACCCAGUUCAGACGAGGUGAGGGCCCUGGAAAAGGGUCUUUCCAGUUCGGCAAAGCUCCUACUGAGCAUACGGAAGAAACUGGCUGCCGUGCCCCACAAGUCUGUCGAGCCA ...(((((........(((((...(((.((((.(((((((......)))))))..(((((..((((.....))))...(....)........))))).)))))))..))))))))))... ( -43.20) >DroSec_CAF1 50485 120 - 1 GAAUUCGAACCCAGUUCAGACGAGGUGAGGGCACUGGAGAAGGGUCUUUCCAGUUCGGCAAAGCUUCUACUAAGCAUCCGAAAGAAAGUGGAUGCCGUGCCCUACAAGUCUGCUGAUCCA ...........(((..(((((...((.(((((((((((((......)))))))).((((...((((.....))))(((((........))))))))).)))))))..))))))))..... ( -42.40) >DroSim_CAF1 53032 120 - 1 GAAUUCGAACCCAGUUCAGACGAGGUGAGGGCACUGGAGAAGGGUCUUUCCAGUUCGGCAAAGCUUCUACUAAGCAUCCGAAAGAAAGUGGAUGCCGUACCCUACAUGUCUGCCGAGCCA .............((((.(..((.(((((((.((((((((......)))))))).((((...((((.....))))(((((........)))))))))..)))).))).))..).)))).. ( -37.50) >DroEre_CAF1 53162 120 - 1 AAAUCCAGUCCCAGUUUGGACGAGAUGCGGGCACUGGAAAAGGGUCUUUCCAGGGCGGCAAAGCUCCUGCUGAGCAUGCGGCAGAAACUGACUGCAGUGGCCUGCAAAUCCGCCAGGCCA ..(((..((((......))))..)))...(((.(((((((......)))))))(((((....((((.....)))).((((((....((((....)))).))).)))...)))))..))). ( -47.90) >DroYak_CAF1 52484 120 - 1 AAAUCCAUACCCAGUUUAGACGAGAUGAGGGCACUGGAAAAGGGACUUUCCAGGUCGGCAAAGCUCCUACUGAGCAUGCGAAAGAAACUGACUGCAGUGCCCUGCAUAUCUGCCAUGCCA ...........((...((((....((((((((((((......((.....)).((((((....((((.....))))...(....)...)))))).))))))))).))).))))...))... ( -36.10) >consensus AAAUUCGAACCCAGUUCAGACGAGGUGAGGGCACUGGAAAAGGGUCUUUCCAGUUCGGCAAAGCUCCUACUGAGCAUCCGAAAGAAACUGGCUGCCGUGCCCUACAAGUCUGCCGAGCCA ................(((((...(((.((((((((((((......)))))))..(((((..((((.....))))...(....)........)))))))))))))..)))))........ (-29.84 = -29.96 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:36:09 2006