
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,117,404 – 25,117,587 |
| Length | 183 |
| Max. P | 0.901755 |

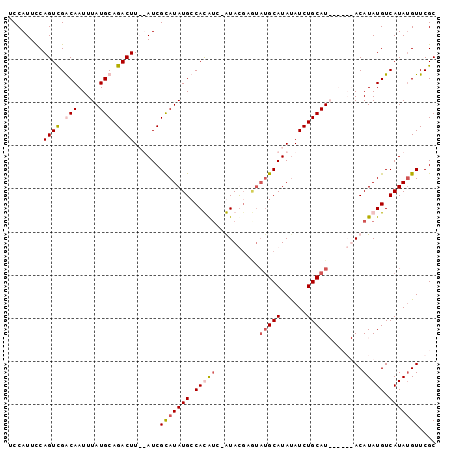
| Location | 25,117,404 – 25,117,494 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -11.78 |
| Energy contribution | -13.22 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25117404 90 + 27905053 UCCAUUUCAGUUGGCAAUUUAUGCAGACUU--AUCGUAUAUGCCACAAC-GUACGAGUAUGCAUAUAUCUGCAU------ACAUAUGUCAUAUGUUCGC .......((..(((((((.((((((((..(--((.((((((........-......)))))))))..)))))))------).)).)))))..))..... ( -20.34) >DroSim_CAF1 48529 90 + 1 UCCAUUCCAGUCGGCAAUUUAUGCAGACUU--AUCGCAUAUGCCACAUC-GUACGAGUAUGCAUAUAUCUGCAU------ACAUAUGUCAUAUGUUCGC ........((((.(((.....))).)))).--...(((((((.((..((-....))(((((((......)))))------))...)).))))))).... ( -22.30) >DroEre_CAF1 48776 99 + 1 UCCAUUCCAGUCGACAAUUUAUGCAGACUUAUAUCGCAUAUGGCAUGUGGAUAUGCAUAUGCAUAUAUCUGCAUACUCGUACAUAUGUCAUAAGUUCGC .........(((((.....((((((((..(((((.(((((((.((((....)))))))))))))))))))))))).))).))................. ( -25.50) >DroYak_CAF1 47893 92 + 1 UCCAUUCCAGUCGACAAUUUAUGCAGACUUAUAUCGCAUAUGCCAUAUC-AUAU------GCAUACAUCUGCAUACUCGUACAUAUGUCAUAUGUUCGC ...................((((((((..(((...(((((((......)-))))------)))))..))))))))..((.((((((...)))))).)). ( -20.80) >consensus UCCAUUCCAGUCGACAAUUUAUGCAGACUU__AUCGCAUAUGCCACAUC_AUACGAGUAUGCAUAUAUCUGCAU______ACAUAUGUCAUAUGUUCGC ........((((.(((.....))).))))......(((((((.(((((..........(((((......)))))........))))).))))))).... (-11.78 = -13.22 + 1.44)


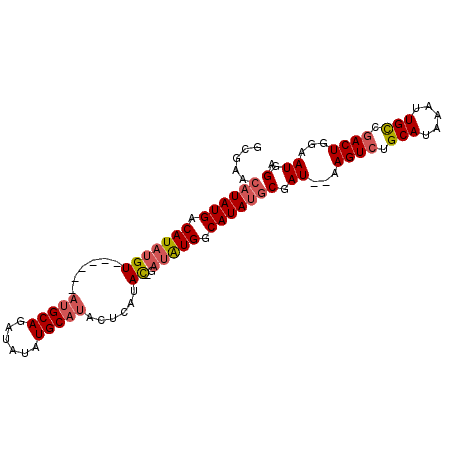
| Location | 25,117,404 – 25,117,494 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25117404 90 - 27905053 GCGAACAUAUGACAUAUGU------AUGCAGAUAUAUGCAUACUCGUAC-GUUGUGGCAUAUACGAU--AAGUCUGCAUAAAUUGCCAACUGAAAUGGA ((((((((((...))))))------((((((((((((((.(((......-...))))))))).....--..))))))))...))))............. ( -20.51) >DroSim_CAF1 48529 90 - 1 GCGAACAUAUGACAUAUGU------AUGCAGAUAUAUGCAUACUCGUAC-GAUGUGGCAUAUGCGAU--AAGUCUGCAUAAAUUGCCGACUGGAAUGGA .....((((((.(((((((------(((((......)))))))......-.))))).))))))....--.((((.(((.....))).))))........ ( -24.41) >DroEre_CAF1 48776 99 - 1 GCGAACUUAUGACAUAUGUACGAGUAUGCAGAUAUAUGCAUAUGCAUAUCCACAUGCCAUAUGCGAUAUAAGUCUGCAUAAAUUGUCGACUGGAAUGGA ......((((..((..((.((((.(((((((((((((((((((((((......))).))))))).))))..)))))))))..))))))..))..)))). ( -27.30) >DroYak_CAF1 47893 92 - 1 GCGAACAUAUGACAUAUGUACGAGUAUGCAGAUGUAUGC------AUAU-GAUAUGGCAUAUGCGAUAUAAGUCUGCAUAAAUUGUCGACUGGAAUGGA .((.((((((...)))))).))..(((((((((((((((------((((-(......))))))).))))..)))))))))................... ( -25.90) >consensus GCGAACAUAUGACAUAUGU______AUGCAGAUAUAUGCAUACUCAUAC_GAUAUGGCAUAUGCGAU__AAGUCUGCAUAAAUUGCCGACUGGAAUGGA .....((((((.(((((((......(((((......)))))......))..))))).))))))(.((...((((.(((.....))).))))...)).). (-15.90 = -16.90 + 1.00)


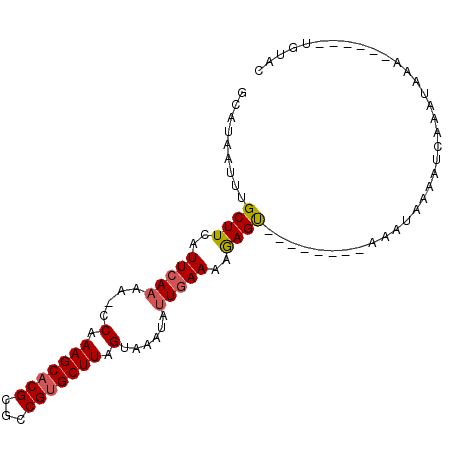
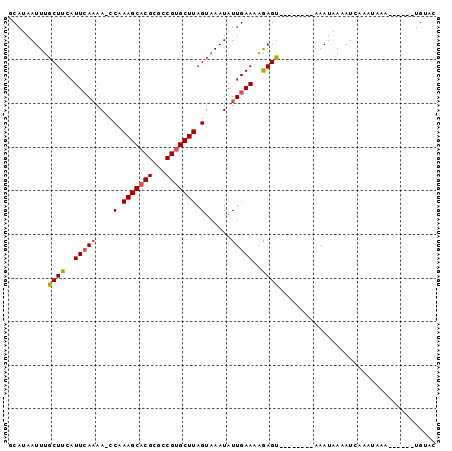
| Location | 25,117,494 – 25,117,587 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -13.93 |
| Consensus MFE | -8.94 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25117494 93 - 27905053 ACAUAAUUCGCUUCAUUCACAAACCAAAGCACGCGCCGGGCUUAGUAAAUAUUGAAAGAAGCUGAAAAUCAAAUAAAAUA-----AAAUGAACUGGUC ......((((((((.((((.......((((.((...)).)))).........)))).))))).)))..............-----............. ( -12.89) >DroVir_CAF1 60716 83 - 1 GCAUAAUUUGCUUCAUUCAAAA-UCCAAGCACGCGCCGUGCUUAGUAAAUAUUGAAAAGAGU--------AAAUAAAAUCAAAUAAA------UGUAC .....((((((((..(((((..-.(.(((((((...))))))).)......)))))..))))--------)))).............------..... ( -16.70) >DroMoj_CAF1 56700 83 - 1 GCAUAAUUUGCUUCAUUGAAAA-CCAAAGCACGCGCCGUGCUUAGUAAAUAUUGAAAAGAGU--------AAAUAAAAUCAAAUAAA------UGUAC .....((((((((((......(-(..(((((((...))))))).))......)))....)))--------)))).............------..... ( -12.20) >consensus GCAUAAUUUGCUUCAUUCAAAA_CCAAAGCACGCGCCGUGCUUAGUAAAUAUUGAAAAGAGU________AAAUAAAAUCAAAUAAA______UGUAC .........((((..(((((....(.(((((((...))))))).)......)))))..)))).................................... ( -8.94 = -9.50 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:36:03 2006