
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,115,489 – 25,115,586 |
| Length | 97 |
| Max. P | 0.978245 |

| Location | 25,115,489 – 25,115,586 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -24.95 |
| Energy contribution | -24.81 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.99 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
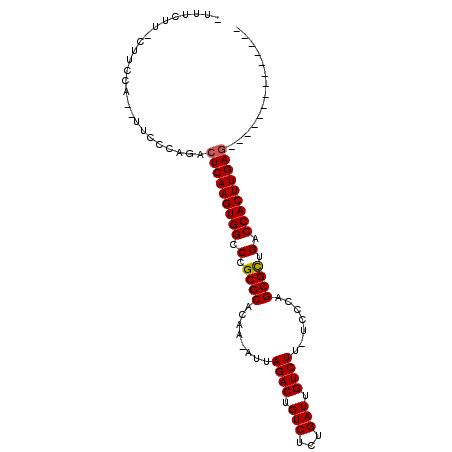
>3R_DroMel_CAF1 25115489 97 + 27905053 CUCGCAUCCUUGGGCUCAAGUGGUCAACGCUGGGA-AAGACAAUCGCAGACAGUCUAAU-UUGUGCGCGGGCCACUUGAGUCUGGGAA--UGGAA--AAGAAA- .((.(((.((..(((((((((((((..(((((.((-.......)).)).((((......-)))))))..)))))))))))))..)).)--)))).--......- ( -40.50) >DroPse_CAF1 49750 86 + 1 --------------CUCAAGUGGUCAGCGCUGGGAAAAGACAAUCAGAGACAGUCUAAU-UUGUGCGCGGGCCACUUGAGCCAUAGAA--AGAAAGAAAGGAC- --------------(((((((((((.((((..(((..((((...........))))..)-))..)))).)))))))))))........--.............- ( -29.50) >DroEre_CAF1 46914 98 + 1 CUCGCAUCCUUGGGCUCAAGUGGUCAACGCUGGGA-AAGACAAUCACAGACAGUCUAAU-UUGUGCGCGGGCCACUUGAGUCUGGGAA--UGGAAA-AAGAAA- .((.(((.((..(((((((((((((..((((..(.-.((((...........))))..)-..).)))..)))))))))))))..)).)--))))..-......- ( -40.40) >DroYak_CAF1 45943 98 + 1 CUGGCAUCCUUGGGCUCAAGUGGUCAACGCUGGGA-AAGACAAUCGCAGACAGUCUAAU-UUGUGCGCGGGCCACUUGAGUCUGGGAA--UGGAAG-AAGAAA- ((..(((.((..(((((((((((((..(((((.((-.......)).)).((((......-)))))))..)))))))))))))..)).)--))..))-......- ( -40.70) >DroAna_CAF1 49156 87 + 1 --------------UUCAAGUGGUCAGCGCUGGGA-AAGACAAUCAGAGACAGUCUAAUUUUGUGCGCGGGCCACUUGAGUCCGGGGAAAAAUAA--AAGAAAU --------------(((((((((((.((((..(((-(((((...........))))..))))..)))).)))))))))))((....)).......--....... ( -27.80) >DroPer_CAF1 49733 86 + 1 --------------CUCAAGUGGUCAGCGCUGGGAAAAGACAAUCAGAGACAGUCUAAU-UUGUGCGCGGGCCACUUGAGCCAUAGAA--AGAAAGAAAGGAC- --------------(((((((((((.((((..(((..((((...........))))..)-))..)))).)))))))))))........--.............- ( -29.50) >consensus ______________CUCAAGUGGUCAACGCUGGGA_AAGACAAUCACAGACAGUCUAAU_UUGUGCGCGGGCCACUUGAGUCUGGGAA__AGGAAG_AAGAAA_ ..............(((((((((((..(((.......((((...........))))........)))..)))))))))))........................ (-24.95 = -24.81 + -0.14)



| Location | 25,115,489 – 25,115,586 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -25.23 |
| Energy contribution | -25.14 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25115489 97 - 27905053 -UUUCUU--UUCCA--UUCCCAGACUCAAGUGGCCCGCGCACAA-AUUAGACUGUCUGCGAUUGUCUU-UCCCAGCGUUGACCACUUGAGCCCAAGGAUGCGAG -......--.((((--(((...(.(((((((((.(.((((....-...((((.(((...))).)))).-.....)))).).))))))))).)...))))).)). ( -30.42) >DroPse_CAF1 49750 86 - 1 -GUCCUUUCUUUCU--UUCUAUGGCUCAAGUGGCCCGCGCACAA-AUUAGACUGUCUCUGAUUGUCUUUUCCCAGCGCUGACCACUUGAG-------------- -.............--........(((((((((.(.((((....-...((((.(((...))).)))).......)))).).)))))))))-------------- ( -26.64) >DroEre_CAF1 46914 98 - 1 -UUUCUU-UUUCCA--UUCCCAGACUCAAGUGGCCCGCGCACAA-AUUAGACUGUCUGUGAUUGUCUU-UCCCAGCGUUGACCACUUGAGCCCAAGGAUGCGAG -......-..((((--(((...(.(((((((((.(.((((....-...((((.(((...))).)))).-.....)))).).))))))))).)...))))).)). ( -30.42) >DroYak_CAF1 45943 98 - 1 -UUUCUU-CUUCCA--UUCCCAGACUCAAGUGGCCCGCGCACAA-AUUAGACUGUCUGCGAUUGUCUU-UCCCAGCGUUGACCACUUGAGCCCAAGGAUGCCAG -......-....((--(((...(.(((((((((.(.((((....-...((((.(((...))).)))).-.....)))).).))))))))).)...))))).... ( -28.72) >DroAna_CAF1 49156 87 - 1 AUUUCUU--UUAUUUUUCCCCGGACUCAAGUGGCCCGCGCACAAAAUUAGACUGUCUCUGAUUGUCUU-UCCCAGCGCUGACCACUUGAA-------------- .......--................((((((((.(.((((........((((.(((...))).)))).-.....)))).).)))))))).-------------- ( -23.74) >DroPer_CAF1 49733 86 - 1 -GUCCUUUCUUUCU--UUCUAUGGCUCAAGUGGCCCGCGCACAA-AUUAGACUGUCUCUGAUUGUCUUUUCCCAGCGCUGACCACUUGAG-------------- -.............--........(((((((((.(.((((....-...((((.(((...))).)))).......)))).).)))))))))-------------- ( -26.64) >consensus _UUUCUU_CUUCCA__UUCCCAGACUCAAGUGGCCCGCGCACAA_AUUAGACUGUCUCUGAUUGUCUU_UCCCAGCGCUGACCACUUGAG______________ ........................(((((((((.(.((((........((((.(((...))).)))).......)))).).))))))))).............. (-25.23 = -25.14 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:36:01 2006