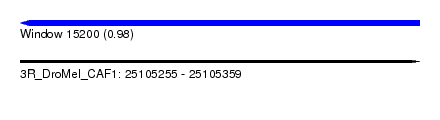
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,105,255 – 25,105,359 |
| Length | 104 |
| Max. P | 0.982937 |

| Location | 25,105,255 – 25,105,359 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 73.66 |
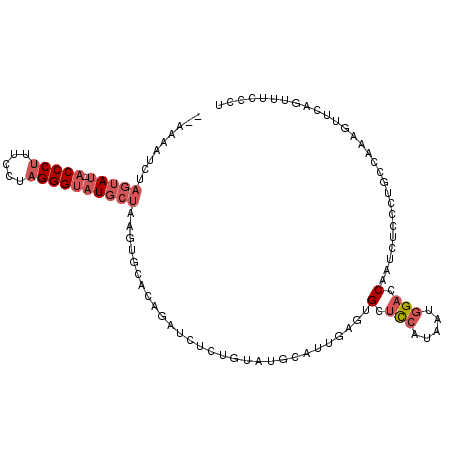
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -7.92 |
| Energy contribution | -9.12 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25105255 104 - 27905053 --AUUAUAGAGUAU-ACCCUUUCCUAGGGUAUGCUAAGUGCACAGAUCUCUGUUUGUAUUGAGUGCUCCAUAAUCGAGCAGUCUCCCUGCCAAAGUUCAUUUUUCCU --.......(((((-(((((.....)))))))))).(((((((((....))))..)))))((.(((((.......))))).))........................ ( -29.30) >DroSec_CAF1 34185 104 - 1 --AGUAUGCAGUAU-ACCCUUUCCUAGGGUAUGCUAAGUGCACAGAUCUCUGUAUGCAUUGAGUGCUCCAUUAUGGCCCAAUCUCCCUGCCAAAGUUCAGCUUCCCU --.((((..(((((-(((((.....))))))))))..))))((((....)))).......(((.(((..(((.((((...........)))).)))..))))))... ( -30.70) >DroSim_CAF1 35146 104 - 1 --AGUAUGCAGUAU-ACCCUUUCCUAGGGUAUGCUAAGUGCACAGAUCUCUGUAUGCAUUGAGUGCUCCAUAAUGGACCAAUCUCCCUGCCAAAGUUCAGCUUCCCU --.((((..(((((-(((((.....))))))))))..))))((((....))))..(((..(((((.(((.....))).))..)))..)))................. ( -31.20) >DroEre_CAF1 34589 92 - 1 GCAAAAUCUAGUAU-ACCCACUCCUAGGGUAUGCUAAGUGCACAGAUCUC--------------GCUCCAUAAUGGAUCAAUCUCCUUGCCAAAGUUCAUUUUCCCU ((((....((((((-((((.......))))))))))........(((((.--------------..........))))).......))))................. ( -20.40) >DroYak_CAF1 35408 104 - 1 --UAAAUCUAGUAU-ACCCUAUCCUAGGGUAUGCUAAGUGCACAGAUCUCUGUAUGCAUUGACUGUUCCAUAAUGGACCAAUCUCCUUGCCAAAGUUCAUUUUUCCU --......((((((-((((((...))))))))))))(((((((((....))))..)))))((.((.(((.....))).)).))........................ ( -28.10) >DroAna_CAF1 39276 91 - 1 --GAAAUCUCAAAAACCCCUUUCAAAGGGUGUCAUAUUU--------------AAGAACUCCCCGUUUCAUAAUGGAGCAAUCUCAUUGCUAAACUUCAGUUCCUCU --............((((((.....)))).)).......--------------..(((((..(((((....)))))((((((...)))))).......))))).... ( -16.50) >consensus __AAAAUCUAGUAU_ACCCUUUCCUAGGGUAUGCUAAGUGCACAGAUCUCUGUAUGCAUUGAGUGCUCCAUAAUGGACCAAUCUCCCUGCCAAAGUUCAGUUUCCCU .........(((((.(((((.....)))))))))).............................(.(((.....))).)............................ ( -7.92 = -9.12 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:35:53 2006