
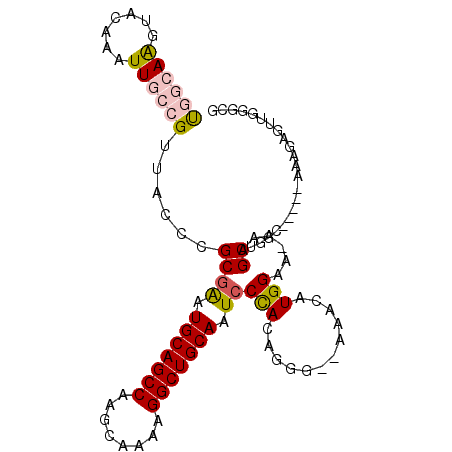
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,099,160 – 25,099,262 |
| Length | 102 |
| Max. P | 0.860661 |

| Location | 25,099,160 – 25,099,262 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.03 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25099160 102 + 27905053 UGGCAAGUACAAAUUGCCGUUACCGGCGAAUGCAGCCAAGCAAAAGGCUGCAAUCCUACAGGGGAAAACAUGGAA-AAAGCUGGCAAAGCAAAGAGUUGGGCG .(((((.......))))).....((((...(((((((........))))))).((((....))))..........-...))))((..(((.....)))..)). ( -32.80) >DroPse_CAF1 32595 89 + 1 CAAGAGCUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGC----GGCUGCAAUCCCA-AGAA--AAACAUGGAAAAAAGCUAAG-----AAAGAGUUA--CC ....((((.....((((........)))).(((((((....----)))))))...(((-.(..--...).))).....))))...-----.........--.. ( -19.80) >DroSec_CAF1 28118 102 + 1 UGGCAAGUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGCAAAAGGCUGCAAUACUACAGGGGAAGCCAUGGAA-AAAGCUGGCAAGGCAAAGAGUUGGGCG ......(..(((.(((((....(((.....(((((((........)))))))........)))...((((.....-.....))))..)))))....)))..). ( -31.52) >DroEre_CAF1 28542 95 + 1 UGGCAAGUACAAAUUGCCGUUACCCGCGGAUGCAGCCAAGCAAAAGGCUGCAAUCCCACUGGG--AAGCAUGGAA-AAAGCUGGC-----AAAGAGUUGGGCG .(((((.......)))))(((.((((.((((((((((........)))))).))))...))))--.)))......-....(..((-----.....))..)... ( -33.90) >DroYak_CAF1 29095 95 + 1 UGGCAAGUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGCAAAAGGCUGCAAUCCCACAGGG--AAACAUGGAA-AAAGCUGCC-----AAAGAGUUGGGCG .(((((.......)))))(((.(((..((.(((((((........))))))).)).....)))--.)))......-...((..((-----((....)))))). ( -31.10) >DroPer_CAF1 32419 88 + 1 CAAGAGCUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGC----GGCUGCAAUCCCA-AGAA--AAACAUGGAA-AAAGCUAAG-----AAAGAGUUA--CC ....((((.....((((........)))).(((((((....----)))))))...(((-.(..--...).)))..-..))))...-----.........--.. ( -19.80) >consensus UGGCAAGUACAAAUUGCCGUUACCCGCGAAUGCAGCCAAGCAAAAGGCUGCAAUCCCACAGGG__AAACAUGGAA_AAAGCUGGC_____AAAGAGUUGGGCG ((((((.......))))))......((((.(((((((........))))))).))(((............)))......))...................... (-17.04 = -17.40 + 0.36)


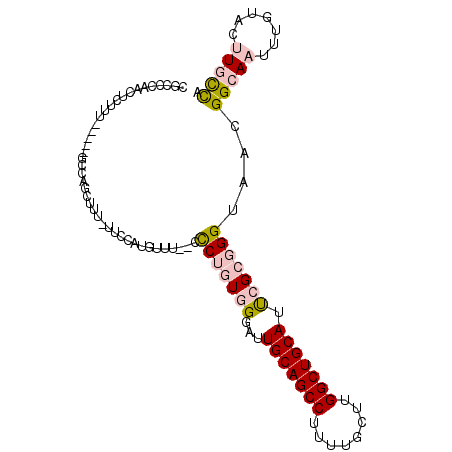
| Location | 25,099,160 – 25,099,262 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.03 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25099160 102 - 27905053 CGCCCAACUCUUUGCUUUGCCAGCUUU-UUCCAUGUUUUCCCCUGUAGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCCGGUAACGGCAAUUUGUACUUGCCA .((...((...((((((((((.((...-..........(((......))).(((((((........)))))))...)).))))).)))))...))....)).. ( -29.60) >DroPse_CAF1 32595 89 - 1 GG--UAACUCUUU-----CUUAGCUUUUUUCCAUGUUU--UUCU-UGGGAUUGCAGCC----GCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUAGCUCUUG ..--.........-----...((((........((((.--..((-((.((.(((((((----....))))))).)).)))).)))).........)))).... ( -25.53) >DroSec_CAF1 28118 102 - 1 CGCCCAACUCUUUGCCUUGCCAGCUUU-UUCCAUGGCUUCCCCUGUAGUAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCA .((...((...(((((..((((.....-.....))))....((((..(...(((((((........))))))).)..))))....)))))...))....)).. ( -32.50) >DroEre_CAF1 28542 95 - 1 CGCCCAACUCUUU-----GCCAGCUUU-UUCCAUGCUU--CCCAGUGGGAUUGCAGCCUUUUGCUUGGCUGCAUCCGCGGGUAACGGCAAUUUGUACUUGCCA .((...((...((-----((((((...-......))).--(((.(((((..(((((((........)))))))))))))))....)))))...))....)).. ( -33.70) >DroYak_CAF1 29095 95 - 1 CGCCCAACUCUUU-----GGCAGCUUU-UUCCAUGUUU--CCCUGUGGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCA .(((.........-----)))......-......(((.--.((((((((..(((((((........))))))))))))))).)))(((((.......))))). ( -33.20) >DroPer_CAF1 32419 88 - 1 GG--UAACUCUUU-----CUUAGCUUU-UUCCAUGUUU--UUCU-UGGGAUUGCAGCC----GCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUAGCUCUUG ..--.........-----...((((..-.....(((..--((((-((.((.(((((((----....))))))).)).)))).))..)))......)))).... ( -25.62) >consensus CGCCCAACUCUUU_____GCCAGCUUU_UUCCAUGUUU__CCCUGUGGGAUUGCAGCCUUUUGCUUGGCUGCAUUCGCGGGUAACGGCAAUUUGUACUUGCCA .........................................(((((((...(((((((........))))))).)))))))....(((((.......))))). (-19.70 = -20.57 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:35:48 2006