
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,096,908 – 25,096,999 |
| Length | 91 |
| Max. P | 0.604564 |

| Location | 25,096,908 – 25,096,999 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -17.43 |
| Energy contribution | -18.93 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
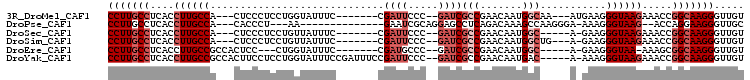
>3R_DroMel_CAF1 25096908 91 - 27905053 CCUUGCCUCACCUUGCCA---CUCCCUCCUGGUAUUUC-------CGAUUCCC--GAUCGCCGAACAAUGGCAA---AUGAAGGGUAAGAAACCGGCAAGGGUUGU ((((((((((..((((((---...((....))......-------(((((...--)))))........))))))---.)))..(((.....))))))))))..... ( -26.90) >DroPse_CAF1 30154 83 - 1 CCUUGCCUCACCUUGCCA---CACCCU---AA--------------GAAUCGCAGGAGCCUCAGACAAAGCCAAGGGA-AAAGGGUAAG--ACCAGGAAGGGUUGC ((((.(((...((((((.---..((((---..--------------(..((..((....))..)).....)..)))).-....))))))--...)))))))..... ( -19.80) >DroSec_CAF1 25847 88 - 1 CCUUGCCUCACCUUGCCA---CUCCCUCCUGUUAUUUC-------CGAUUCCC--GAUCGCCGAACAAUGGC-----A-GAAGGGUAAGAAACCGGCAAGGGUUGU (((((((....((((...---..((((.((((((((((-------(((((...--)))))..))..))))))-----)-).)))))))).....)))))))..... ( -27.60) >DroSim_CAF1 26789 90 - 1 CCUUGCCUCACCUUGCCA---CUCCCUCCUGUUAUUUC-------CGAUUCCC--GAUCGCCGAACAAUGGCUG---A-GAAGGGUAAGAAACCGGCAAGGGUUGU (((((((....((((((.---((..(((..((((((((-------(((((...--)))))..))..)))))).)---)-).)))))))).....)))))))..... ( -28.40) >DroEre_CAF1 26301 87 - 1 CCUUGCCUCACCUUGCCGCCACUCC---CUGGUAUUUC-------CGAUGCCC--GAUCGCCGAACAAUGGC-----A-GAAGGGUAA-AAAGCGGCAAGGGUUGU ..........(((((((((....((---((((((((..-------.)))))).--....((((.....))))-----.-..))))...-...)))))))))..... ( -32.60) >DroYak_CAF1 26811 98 - 1 CCUUGCCUCACCUUGCCGCCACUUCCUCCUGGUAUUUCCGAUUUCCGAUUCCC--GAUCGCCGAACAAUGAC-----A-AAAGGGUAAGAAACCGGCAAGGGUUGU (((((((....((((((((((........))))............(((((...--)))))............-----.-....)))))).....)))))))..... ( -25.10) >consensus CCUUGCCUCACCUUGCCA___CUCCCUCCUGGUAUUUC_______CGAUUCCC__GAUCGCCGAACAAUGGC_____A_GAAGGGUAAGAAACCGGCAAGGGUUGU (((((((....((((((.............................((((.....))))(((.......)))...........)))))).....)))))))..... (-17.43 = -18.93 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:35:47 2006