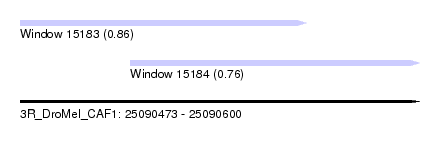
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,090,473 – 25,090,600 |
| Length | 127 |
| Max. P | 0.862256 |

| Location | 25,090,473 – 25,090,564 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.68 |
| Mean single sequence MFE | -40.88 |
| Consensus MFE | -29.51 |
| Energy contribution | -30.36 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25090473 91 + 27905053 UGUAUGCGCCAGCAGAUCCUUUUGUCAGGCCAAAUGCGCGAAAG-UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCU----U-------------CAGCUA (((.((((((.((.(((......)))..(((....((((....)-)))))))).))))))))).(((((.......)))))(((.----.-------------..))). ( -35.70) >DroSec_CAF1 19534 105 + 1 UGUAUGCGCCAGCAGUUCCUUUGGUCAGCCCAAAUGCGCGAAAGAUGUGGCGCAGGCGCAGCAGGAUCCUGGAUAUGGAUCAGCU----UCAGGCCCGGGAUUCAGCUA .....(((((.........(((((.....)))))(((((.(......).))))))))))(((.(((((((((...((((.....)----)))...))))))))).))). ( -41.40) >DroSim_CAF1 19924 104 + 1 UGUAUGCGCCAGCAGAUCCUUUUGUCAGCCCAAAUGCGCGAAAG-UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCU----UCAGGCUCGGGAUUCAGCUA .....(((((.((.(((......))).(((.....((((....)-)))))))).)))))(((.((((((((..((((((.....)----)))))..)))))))).))). ( -43.90) >DroEre_CAF1 19920 107 + 1 UGUAUGCGCCAGCAGAUCCUUUUGUCAGCCCAAAUGCGCGAAAG-UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCUCGCUUCCAACUCG-GAUUCAACUG (((.((((((.((.(((......))).(((.....((((....)-)))))))).)))))))))((((((..((.(((((..((....))))))).)))-)))))..... ( -39.60) >DroYak_CAF1 20179 107 + 1 UGUAUGCGCCAGCAGAUCCUUUUGUCAGCCCAAAUGCGCGAAAG-UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCUCGGUUCAAACUCG-GAUUCAGCUA .....(((((.((.(((......))).(((.....((((....)-)))))))).)))))(((.((((((.(..((((((((.....))))))))..))-))))).))). ( -43.80) >consensus UGUAUGCGCCAGCAGAUCCUUUUGUCAGCCCAAAUGCGCGAAAG_UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCU____UCAAACUCG_GAUUCAGCUA (((.((((((.(((((....))))).........(((((.(......).)))))))))))))).(((((.......)))))((((....((........))...)))). (-29.51 = -30.36 + 0.85)


| Location | 25,090,508 – 25,090,600 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -39.86 |
| Consensus MFE | -30.07 |
| Energy contribution | -31.76 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25090508 92 + 27905053 GCGCGAAAG-UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCU----U-------------CAGCUACAAAAUCAGGACACUGCGCACGUGCGUUGGG---AAG-UC ((((....)-)))((((((.(.((.((((..((((((.((((....(((.----.-------------..))).)))).))))))..)))))).).))))))...---...-.. ( -40.90) >DroSec_CAF1 19569 106 + 1 GCGCGAAAGAUGUGGCGCAGGCGCAGCAGGAUCCUGGAUAUGGAUCAGCU----UCAGGCCCGGGAUUCAGCUACAAAAUCAGGACACUGCGCACGUGCGUUGGG---AAG-UC ((((....).)))((((((.(.(((((.(((((((((...((((.....)----)))...))))))))).))).......(((....))).)).).))))))...---...-.. ( -36.70) >DroSim_CAF1 19959 105 + 1 GCGCGAAAG-UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCU----UCAGGCUCGGGAUUCAGCUACAAAAUCAGGACACUGCGCACGUGCGUUGGG---AAG-UC ((((....)-)))((((((.(.((.((((..((((((.((((((......----)).((((........)))).)))).))))))..)))))).).))))))...---...-.. ( -41.80) >DroEre_CAF1 19955 102 + 1 GCGCGAAAG-UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCUCGCUUCCAACUCG-GAUUCAACUGCGAAAUCAGGAC-------ACGUGCGUUGGG---AAGUUU ((((....)-)))((((((.((((((..((((((..((.(((((..((....))))))).)))-)))))..)))))..........-------.).))))))...---...... ( -33.70) >DroYak_CAF1 20214 112 + 1 GCGCGAAAG-UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCUCGGUUCAAACUCG-GAUUCAGCUACAAAAUCAGGACACUGCGCACGUGCGUUGGGAGGAAGUUU ((((....)-)))((((((.(.((.((((..((((((.((((....((((..((((......)-)))..)))).)))).))))))..)))))).).))))))............ ( -46.20) >consensus GCGCGAAAG_UGUGGCGCAGGCGCAGCAGGAUCCUGGAUUUGGAUCAGCU____UCAAACUCG_GAUUCAGCUACAAAAUCAGGACACUGCGCACGUGCGUUGGG___AAG_UC ((((....).)))((((((.(.((.((((..((((((.((((....((((....((........))...)))).)))).))))))..)))))).).))))))............ (-30.07 = -31.76 + 1.69)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:35:39 2006