
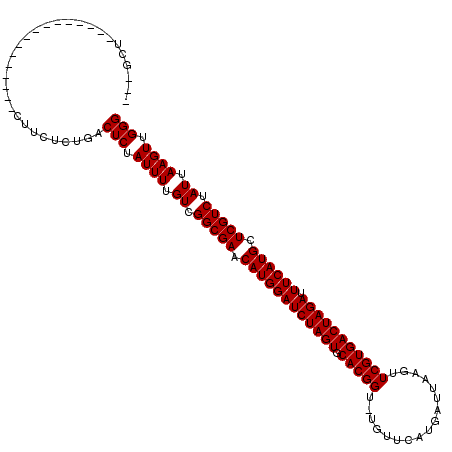
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,042,898 – 25,043,016 |
| Length | 118 |
| Max. P | 0.999729 |

| Location | 25,042,898 – 25,042,998 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -28.53 |
| Energy contribution | -28.53 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25042898 100 + 27905053 ---GCU----------------CUUUUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGU-GGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGG ---...----------------(((..(((((........)))(((((.((((((((((((.(((((.-...............))))))))))).)))))).))))).....))..))) ( -28.69) >DroPse_CAF1 63871 100 + 1 ---GCU----------------CUUCUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGU-UGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGG ---.((----------------(..((..(((........)))(((((.((((((((((((.(((((.-...............))))))))))).)))))).))))).....))..))) ( -28.89) >DroWil_CAF1 71374 120 + 1 GGAGUCCUUUCUAUGUGGCAGCGUCUCUUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGUUUGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGG ((((((......((((....)))).....))))))........(((((.((((((((((((.(((((.................))))))))))).)))))).)))))............ ( -33.63) >DroMoj_CAF1 121730 100 + 1 ---GCU----------------CUUCUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGU-UGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGG ---.((----------------(..((..(((........)))(((((.((((((((((((.(((((.-...............))))))))))).)))))).))))).....))..))) ( -28.89) >DroAna_CAF1 70357 103 + 1 GGGGUC----------------UUUUUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGU-GUUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGG ((((((----------------.......))))))........(((((.((((((((((((.(((((.-.((........))..))))))))))).)))))).)))))............ ( -32.20) >DroPer_CAF1 73472 100 + 1 ---GCU----------------CUUCUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGU-UGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGG ---.((----------------(..((..(((........)))(((((.((((((((((((.(((((.-...............))))))))))).)))))).))))).....))..))) ( -28.89) >consensus ___GCU________________CUUCUCUGACUCUAUUUUGUCGGCGAACAUGGAUCUAGUGCACGGU_UGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGG ...............................(((.((((.((.(((((.((((((((((((.(((((.................))))))))))).)))))).))))).)).)))).))) (-28.53 = -28.53 + -0.00)


| Location | 25,042,898 – 25,042,998 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -24.91 |
| Energy contribution | -24.91 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.96 |
| SVM RNA-class probability | 0.999729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25042898 100 - 27905053 CCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACC-ACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAAAAG----------------AGC--- .....(((....(.((((((((..(((((((((((................-..)))).))))))).)))))))).)(((........)))....)))----------------...--- ( -25.47) >DroPse_CAF1 63871 100 - 1 CCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACA-ACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAGAAG----------------AGC--- .....(((....(.((((((((..(((((((((((................-..)))).))))))).)))))))).)(((........))).)))...----------------...--- ( -25.87) >DroWil_CAF1 71374 120 - 1 CCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACAAACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAAGAGACGCUGCCACAUAGAAAGGACUCC .((.........(.((((((((..(((((((((((...................)))).))))))).)))))))).).......(((.(((....))).)))...........))..... ( -28.41) >DroMoj_CAF1 121730 100 - 1 CCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACA-ACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAGAAG----------------AGC--- .....(((....(.((((((((..(((((((((((................-..)))).))))))).)))))))).)(((........))).)))...----------------...--- ( -25.87) >DroAna_CAF1 70357 103 - 1 CCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAAC-ACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAAAAA----------------GACCCC .....(((....(.((((((((..(((((((((((................-..)))).))))))).)))))))).)(((........))).....))----------------)..... ( -25.47) >DroPer_CAF1 73472 100 - 1 CCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACA-ACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAGAAG----------------AGC--- .....(((....(.((((((((..(((((((((((................-..)))).))))))).)))))))).)(((........))).)))...----------------...--- ( -25.87) >consensus CCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACA_ACCGUGCACUAGAUCCAUGUUCGCCGACAAAAUAGAGUCAGAAAAG________________AGC___ ............(.((((((((..(((((((((((...................)))).))))))).)))))))).)(((........)))............................. (-24.91 = -24.91 + 0.00)



| Location | 25,042,919 – 25,043,016 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -26.95 |
| Energy contribution | -26.56 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25042919 97 + 27905053 GUCGGCGAACAUGGAUCUAGUGCACGGU-GGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACA------------------ACGAAGAGAG .(((((((.((((((((((((.(((((.-...............))))))))))).)))))).)))))......((((........))------------------))))...... ( -29.79) >DroVir_CAF1 89414 114 + 1 GUCGGCGAACAUGGAUCUAGUGCACGGU-UGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAACACACACACACACACACAUAAAAGAAAAACA- .(((((((.((((((((((((.(((((.-...............))))))))))).)))))).)))))......((((...))))......................))......- ( -27.19) >DroGri_CAF1 73634 98 + 1 GUCGGCGAACAUGGAUCUAGUGCACGGU-UGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAACACACAAC--------------ACAAAAA--- ...(((((.((((((((((((.(((((.-...............))))))))))).)))))).)))))......((((.((....)).))))--------------.......--- ( -28.99) >DroWil_CAF1 71414 95 + 1 GUCGGCGAACAUGGAUCUAGUGCACGGUUUGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACA------------------ACAA-GAG-- .(((((((.((((((((((((.(((((.................))))))))))).)))))).)))))......((((........))------------------))..-)).-- ( -29.63) >DroMoj_CAF1 121751 100 + 1 GUCGGCGAACAUGGAUCUAGUGCACGGU-UGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAAUACACACA--------------GCGAAAACA- .(((((((.((((((((((((.(((((.-...............))))))))))).)))))).)))))......((((.((......)).))--------------)))).....- ( -29.49) >DroAna_CAF1 70381 96 + 1 GUCGGCGAACAUGGAUCUAGUGCACGGU-GUUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAGCACA------------------CCAAAGAGU- .(((((((.((((((((((((.(((((.-.((........))..))))))))))).)))))).))))).......((((((....)).------------------)))).))..- ( -28.80) >consensus GUCGGCGAACAUGGAUCUAGUGCACGGU_UGUUCAUGAUUAAGUUCGUGACUAGAUUUCAUGCUCGUCUAUUAAGUUGGGUCAACACA__________________ACAAAAAGA_ ...(((((.((((((((((((.(((((.................))))))))))).)))))).)))))......((((...))))............................... (-26.95 = -26.56 + -0.39)



| Location | 25,042,919 – 25,043,016 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -22.41 |
| Energy contribution | -22.41 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25042919 97 - 27905053 CUCUCUUCGU------------------UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACC-ACCGUGCACUAGAUCCAUGUUCGCCGAC ........((------------------((........))))......(.((((((((..(((((((((((................-..)))).))))))).)))))))).)... ( -24.57) >DroVir_CAF1 89414 114 - 1 -UGUUUUUCUUUUAUGUGUGUGUGUGUGUGUGUUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACA-ACCGUGCACUAGAUCCAUGUUCGCCGAC -......................((.(((..((((...))))...))).))(((((((..(((((((((((................-..)))).))))))).)))))))...... ( -25.57) >DroGri_CAF1 73634 98 - 1 ---UUUUUGU--------------GUUGUGUGUUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACA-ACCGUGCACUAGAUCCAUGUUCGCCGAC ---.......--------------((((.((....)).))))......(.((((((((..(((((((((((................-..)))).))))))).)))))))).)... ( -25.67) >DroWil_CAF1 71414 95 - 1 --CUC-UUGU------------------UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACAAACCGUGCACUAGAUCCAUGUUCGCCGAC --...-..((------------------((........))))......(.((((((((..(((((((((((...................)))).))))))).)))))))).)... ( -24.21) >DroMoj_CAF1 121751 100 - 1 -UGUUUUCGC--------------UGUGUGUAUUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACA-ACCGUGCACUAGAUCCAUGUUCGCCGAC -.....((((--------------...((.((((((......)))))).))(((((((..(((((((((((................-..)))).))))))).)))))))).))). ( -26.77) >DroAna_CAF1 70381 96 - 1 -ACUCUUUGG------------------UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAAAC-ACCGUGCACUAGAUCCAUGUUCGCCGAC -.....((((------------------.((....)))))).......(.((((((((..(((((((((((................-..)))).))))))).)))))))).)... ( -25.77) >consensus _UCUCUUUGU__________________UGUGCUGACCCAACUUAAUAGACGAGCAUGAAAUCUAGUCACGAACUUAAUCAUGAACA_ACCGUGCACUAGAUCCAUGUUCGCCGAC ................................................(.((((((((..(((((((((((...................)))).))))))).)))))))).)... (-22.41 = -22.41 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:35:09 2006