
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,039,465 – 25,039,565 |
| Length | 100 |
| Max. P | 0.922768 |

| Location | 25,039,465 – 25,039,565 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -18.37 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
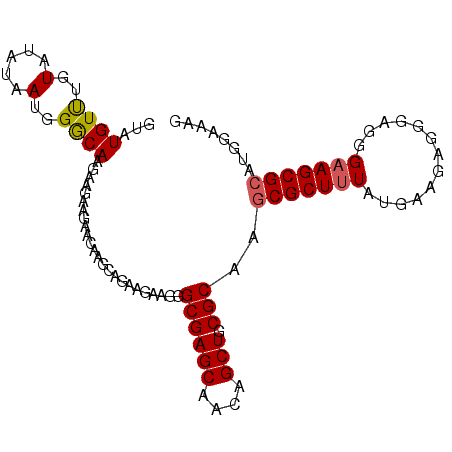
>3R_DroMel_CAF1 25039465 100 + 27905053 GUAUGUUUGUAUAUAAUGGACAAGAAGAAGAAUAAGCAGAAGAAGGGCGAGCAACAGCUGCGCAAGCGCUUUAUGAAGAAUGAGGGUAGCGCAAGGAAAG ...((((..(.....)..)))).............((.........((((((....))).)))..((.((((((.....))))))...))))........ ( -19.40) >DroSec_CAF1 60225 100 + 1 GUAUGUUUGUAUAUAAUGGGCAAGAAGAAGAACAAGCAGAAGAAGGGCGAGCAACAGCUGCGCAAGCGCUUUAUGAAUAGGGAGGGAAGCGCAUGGAAAG ...(((((((.....................)))))))........((((((....))).)))..(((((((.............)))))))........ ( -20.92) >DroSim_CAF1 54607 100 + 1 GUAUGUUUGUAUAUAAUGGGCAAGAAGAAGAACAAGCAGAAGAAGGGCGAGCAACAGCUGCGCAAGCGCUUUAAGAAUAGGGAGGGAAGCGAAUGGAAAG ...(((((((.....................)))))))........((((((....))).)))...((((((.............))))))......... ( -16.92) >DroEre_CAF1 62671 92 + 1 -UAUGUCUGUAUAUAAUGGGCAAGAAGA------AGCAGCUGAAGGGCGAGCAACAGCUGCGCAAGCGCUUUGCAAAGAGGGUGCGAAGCGCAGGG-AAG -..(((((((.....)))))))......------.(((((((..(......)..)))))))....((((((((((.......))))))))))....-... ( -34.10) >consensus GUAUGUUUGUAUAUAAUGGGCAAGAAGAAGAACAAGCAGAAGAAGGGCGAGCAACAGCUGCGCAAGCGCUUUAUGAAGAGGGAGGGAAGCGCAUGGAAAG ...((((..(.....)..))))........................((((((....))).)))..(((((((.............)))))))........ (-18.37 = -18.50 + 0.12)

| Location | 25,039,465 – 25,039,565 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -15.26 |
| Consensus MFE | -11.71 |
| Energy contribution | -12.21 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25039465 100 - 27905053 CUUUCCUUGCGCUACCCUCAUUCUUCAUAAAGCGCUUGCGCAGCUGUUGCUCGCCCUUCUUCUGCUUAUUCUUCUUCUUGUCCAUUAUAUACAAACAUAC ........(((((.................)))))..(((.(((....)))))).............................................. ( -13.03) >DroSec_CAF1 60225 100 - 1 CUUUCCAUGCGCUUCCCUCCCUAUUCAUAAAGCGCUUGCGCAGCUGUUGCUCGCCCUUCUUCUGCUUGUUCUUCUUCUUGCCCAUUAUAUACAAACAUAC ........((((((...............))))))..(((.(((....)))))).........((..(........)..))................... ( -14.26) >DroSim_CAF1 54607 100 - 1 CUUUCCAUUCGCUUCCCUCCCUAUUCUUAAAGCGCUUGCGCAGCUGUUGCUCGCCCUUCUUCUGCUUGUUCUUCUUCUUGCCCAUUAUAUACAAACAUAC .........(((((...............)))))...(((.(((....)))))).........((..(........)..))................... ( -10.16) >DroEre_CAF1 62671 92 - 1 CUU-CCCUGCGCUUCGCACCCUCUUUGCAAAGCGCUUGCGCAGCUGUUGCUCGCCCUUCAGCUGCU------UCUUCUUGCCCAUUAUAUACAGACAUA- ...-....((((((.(((.......))).))))))....(((((((............))))))).------...........................- ( -23.60) >consensus CUUUCCAUGCGCUUCCCUCCCUAUUCAUAAAGCGCUUGCGCAGCUGUUGCUCGCCCUUCUUCUGCUUGUUCUUCUUCUUGCCCAUUAUAUACAAACAUAC ........((((((...............))))))..(((.(((....)))))).............................................. (-11.71 = -12.21 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:35:02 2006