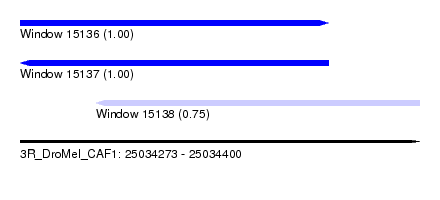
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,034,273 – 25,034,400 |
| Length | 127 |
| Max. P | 0.999688 |

| Location | 25,034,273 – 25,034,371 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -29.53 |
| Energy contribution | -29.87 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25034273 98 + 27905053 UUUAUCGCUCCAGCUCCUUCAACUUGGACGCCAUCAUCUGCAGAAGCAGCUGAAGCCCCAGCUGAAGCUCCAGCUGAAGCUCCAGCUGGCAGCGGUGA .((((((((((((((((........))).....(((.((((....)))).)))(((..((((((......))))))..)))...))))).)))))))) ( -40.70) >DroSim_CAF1 49562 83 + 1 UUUAUCGCACCAGCUC---CAACUCGGACGCCAUCAUCUGCAGAAGCAGCUGAA------------GCUCCAGCUGAAGCCCCAGCUGGCAGCGGUGA .......((((.((((---(.....))).))..(((.((((....)))).))).------------((((((((((......))))))).))))))). ( -34.90) >DroEre_CAF1 57391 83 + 1 GCUGAUGCUCCAACUC---CAAGUUGGACGCCAUCAUCUCCAGAACCAGCUGAA------------GCCCCAGCUGAAGCUCCAGCUGGCGGCGGUGG ((....))((((((..---...))))))((((........(((......)))..------------((((((((((......))))))).))))))). ( -35.30) >consensus UUUAUCGCUCCAGCUC___CAACUUGGACGCCAUCAUCUGCAGAAGCAGCUGAA____________GCUCCAGCUGAAGCUCCAGCUGGCAGCGGUGA .........((((..........)))).((((.(((.((((....)))).))).............((((((((((......))))))).))))))). (-29.53 = -29.87 + 0.34)


| Location | 25,034,273 – 25,034,371 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -44.17 |
| Consensus MFE | -25.97 |
| Energy contribution | -25.20 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25034273 98 - 27905053 UCACCGCUGCCAGCUGGAGCUUCAGCUGGAGCUUCAGCUGGGGCUUCAGCUGCUUCUGCAGAUGAUGGCGUCCAAGUUGAAGGAGCUGGAGCGAUAAA .........((((((((((((((....)))))))))))))).(((((((((.((((.((.((((....))))...)).)))).)))))))))...... ( -51.70) >DroSim_CAF1 49562 83 - 1 UCACCGCUGCCAGCUGGGGCUUCAGCUGGAGC------------UUCAGCUGCUUCUGCAGAUGAUGGCGUCCGAGUUG---GAGCUGGUGCGAUAAA .(((((((.((((((.(((((((....)))((------------((((.((((....)))).))).))))))).)))))---)))).))))....... ( -37.60) >DroEre_CAF1 57391 83 - 1 CCACCGCCGCCAGCUGGAGCUUCAGCUGGGGC------------UUCAGCUGGUUCUGGAGAUGAUGGCGUCCAACUUG---GAGUUGGAGCAUCAGC ...(((..(((((((((((((((....)))))------------))))))))))..)))...(((((.(.((((((...---..)))))))))))).. ( -43.20) >consensus UCACCGCUGCCAGCUGGAGCUUCAGCUGGAGC____________UUCAGCUGCUUCUGCAGAUGAUGGCGUCCAAGUUG___GAGCUGGAGCGAUAAA .....(((.((((((((....)))))))))))............(((((((......((.((((....))))...))......)))))))........ (-25.97 = -25.20 + -0.77)

| Location | 25,034,297 – 25,034,400 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -17.81 |
| Energy contribution | -19.28 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25034297 103 - 27905053 CCUGG------UACUUAUUGAUCACUUCAGGCCUUUCACCGCUGCCAGCUGGAGCUUCAGCU-GGAGCUUCAGCUGGGGCUUCAGCUGCUUCUGCAGAUGAUGGCGUCCA ...((------.((...((((.....))))(((..(((...(((((((((((((((((((((-((....))))))))))))))))))).....)))).))).))))))). ( -48.30) >DroSec_CAF1 55226 79 - 1 CCUGG------UACUUAUUGAUCACUUCAGGCCUUUCACCGCUGCCAGC------------U-GGAGC------------UUCAGCUGCUUCUGCAGAUGAUGGCGUCCG ...((------.((...((((.....))))(((..(((...((((((((------------(-((...------------.))))))).....)))).))).))))))). ( -24.10) >DroSim_CAF1 49583 91 - 1 CCUGG------UACUUAUUGAUCACUUCAGGCCUUUCACCGCUGCCAGCUGGGGCUUCAGCU-GGAGC------------UUCAGCUGCUUCUGCAGAUGAUGGCGUCCG ...((------.((...((((.....))))(((..(((...(((((((((((((((((....-)))))------------)))))))).....)))).))).))))))). ( -36.50) >DroEre_CAF1 57412 91 - 1 CCUGG------UACUUAUUGAUCACUUCAGGCCUUCCACCGCCGCCAGCUGGAGCUUCAGCU-GGGGC------------UUCAGCUGGUUCUGGAGAUGAUGGCGUCCA ...((------.((....(.((((((((((((........)))(((((((((((((((....-)))))------------))))))))))..))))).)))).).)))). ( -39.00) >DroYak_CAF1 50572 98 - 1 CCUGGUACUGGUACUUAUUGAUCACUUCAGGCCUUUCACCACCGCCAGCCGGAGCUUCAGCUUUGGGC------------UUCAGCUGGUUCUGCAAAUGAUGGCGUCCA .((((...((((........))))..))))(((..(((.((..((((((.(((((((.......))))------------))).))))))..))....))).)))..... ( -28.80) >consensus CCUGG______UACUUAUUGAUCACUUCAGGCCUUUCACCGCUGCCAGCUGGAGCUUCAGCU_GGAGC____________UUCAGCUGCUUCUGCAGAUGAUGGCGUCCA ...((............((((.....))))(((.......(((.(.((((((....)))))).).))).............(((.((((....)))).))).)))..)). (-17.81 = -19.28 + 1.47)


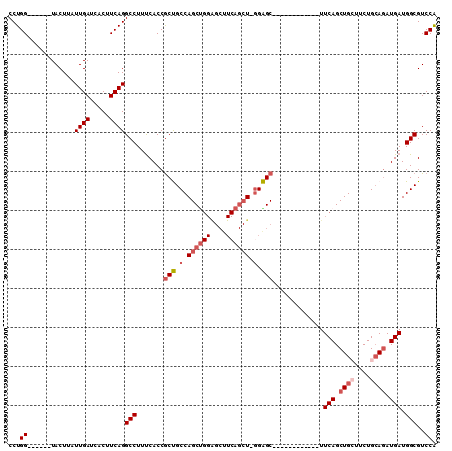
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:58 2006