
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,018,254 – 25,018,349 |
| Length | 95 |
| Max. P | 0.967583 |

| Location | 25,018,254 – 25,018,349 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -14.08 |
| Energy contribution | -15.96 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
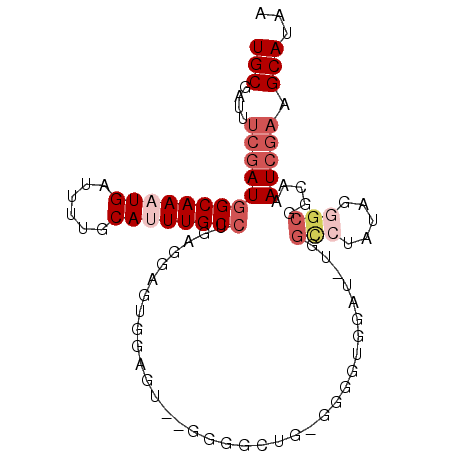
>3R_DroMel_CAF1 25018254 95 + 27905053 UGCGAUUUCGAUGGCAAAUGAUUUUGCAUUUGCCUGAGCGGUGGCGU--GGGGCUGUGGGGUGGAUAUGGCGUAUAGGGCGGCAAAUCGAAGCAUAA (((...((((((((((((((......))))))))......((.((.(--.(.((((((.......)))))).)...).)).))..)))))))))... ( -29.20) >DroSec_CAF1 39434 96 + 1 UGCGAUUUCGAUGGCAAAUGAUUUUGCAUUUGCCUGAGCUGUGGAGUGUGGGGCUG-GGGGUGGAUUUGGCCCAUAGGGCGGCAAAUCGAAGCAUAA (((...((((((((((((((......))))))))...(((((....((((((.(..-((......))..)))))))..)))))..)))))))))... ( -35.00) >DroSim_CAF1 33571 94 + 1 UGCGAUUUCGAUGGCAAAUGAUUUUGCAUUUGCCGGAGGGGUGGAGU--GGGCCUG-GGUGUGGAUUUGGCCUAUAGGGCGGCAAAUCGAAGCAUAA (((.((..(..(((((((((......)))))))))..)..))...((--.(.((((-(((.........))))..))).).))........)))... ( -30.60) >DroEre_CAF1 41447 91 + 1 UGCGAUUUCGAUGGCAAAUGAUUUUGCAGUUGCCUGAAGAGCGGAGA--GGGUAUGGGGGGCG----UGGUCUGUGGGGCGGCAAAUCGAAGCAUAA (((...((((((.(((((....))))).(((((((.....(((((..--(.((.......)).----)..))))).)))))))..)))))))))... ( -27.90) >DroAna_CAF1 44282 79 + 1 UGCGAUUUGGAUGGCAAAUGAUUUUGCAGUUGCAA--GGAACGGGCG-----GU---UGCACGGAU--------CAGGGCGGUAAAUGGUAGCAUAA (((.(((...((.((...(((((((((((..((..--.......)).-----.)---)))).))))--------))..)).))....))).)))... ( -21.50) >consensus UGCGAUUUCGAUGGCAAAUGAUUUUGCAUUUGCCUGAGGAGUGGAGU__GGGGCUG_GGGGUGGAU_UGGCCUAUAGGGCGGCAAAUCGAAGCAUAA (((....(((((((((((((......))))))))...................................(((.....))).....))))).)))... (-14.08 = -15.96 + 1.88)


| Location | 25,018,254 – 25,018,349 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -17.10 |
| Consensus MFE | -10.68 |
| Energy contribution | -12.88 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25018254 95 - 27905053 UUAUGCUUCGAUUUGCCGCCCUAUACGCCAUAUCCACCCCACAGCCCC--ACGCCACCGCUCAGGCAAAUGCAAAAUCAUUUGCCAUCGAAAUCGCA ...(((((((((..((.((.......))...............((...--..))....))...((((((((......))))))))))))))...))) ( -17.20) >DroSec_CAF1 39434 96 - 1 UUAUGCUUCGAUUUGCCGCCCUAUGGGCCAAAUCCACCCC-CAGCCCCACACUCCACAGCUCAGGCAAAUGCAAAAUCAUUUGCCAUCGAAAUCGCA ...((((((((((((..((((...))))))))))......-......................((((((((......))))))))...)))...))) ( -23.10) >DroSim_CAF1 33571 94 - 1 UUAUGCUUCGAUUUGCCGCCCUAUAGGCCAAAUCCACACC-CAGGCCC--ACUCCACCCCUCCGGCAAAUGCAAAAUCAUUUGCCAUCGAAAUCGCA ...(((((((((.............((((...........-..)))).--.............((((((((......))))))))))))))...))) ( -22.72) >DroEre_CAF1 41447 91 - 1 UUAUGCUUCGAUUUGCCGCCCCACAGACCA----CGCCCCCCAUACCC--UCUCCGCUCUUCAGGCAACUGCAAAAUCAUUUGCCAUCGAAAUCGCA ...((((((((((((((.............----..............--.............)))))..(((((....))))).))))))...))) ( -13.00) >DroAna_CAF1 44282 79 - 1 UUAUGCUACCAUUUACCGCCCUG--------AUCCGUGCA---AC-----CGCCCGUUCC--UUGCAACUGCAAAAUCAUUUGCCAUCCAAAUCGCA ...(((...........((...(--------(..((.((.---..-----.)).)).)).--..))....(((((....)))))..........))) ( -9.50) >consensus UUAUGCUUCGAUUUGCCGCCCUAUAGGCCA_AUCCACCCC_CAGCCCC__ACUCCACCCCUCAGGCAAAUGCAAAAUCAUUUGCCAUCGAAAUCGCA ...(((((((((.....(((.....)))...................................((((((((......))))))))))))))...))) (-10.68 = -12.88 + 2.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:51 2006