
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,008,542 – 25,008,691 |
| Length | 149 |
| Max. P | 0.958487 |

| Location | 25,008,542 – 25,008,651 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -19.06 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
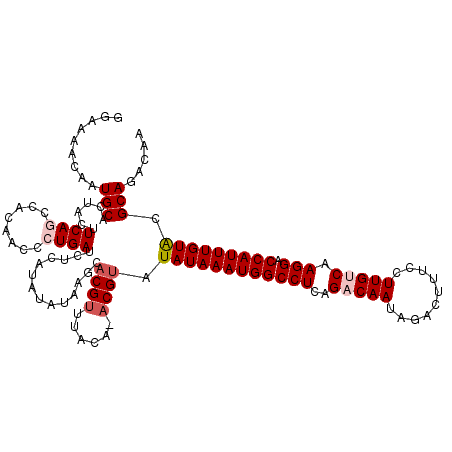
>3R_DroMel_CAF1 25008542 109 - 27905053 AUAUAAGCACGUUUACA-ACGUAUAUAAAUGGCCUCAGACAAUAGACUCUCCUUGUCAAGGACCAUUUGUAAGCAGACAAAUUUCUAAAAAUACUUGCGGUUGACAUUCU ...............((-((((.((((((((((((..(((((..........))))).))).))))))))).))(((......))).............))))....... ( -22.00) >DroSec_CAF1 30970 110 - 1 AUAUAAGCACGUUUACAAACGUAUAUAAAUGGCCUCAGACAAUAGACUUUCCUUGUCAAGGACCAUUUGUAUGCAGACGAAUUUCUAAAAAUACUUGCGGUUGACAUUCU ......(((.((........(((((((((((((((..(((((..........))))).))).))))))))))))(((......)))......)).)))............ ( -25.60) >DroSim_CAF1 25084 110 - 1 AUAUAAGCACGCUUACAAACGUAUAUAAAUGGCCUCAGACAAUAGACUUUCCUUGUCAAGGACCAUUUGUAUGCAGACAAAUUUCUAAAAAUACCUGCGGUUGACAUUCU .....(((.(((........(((((((((((((((..(((((..........))))).))).))))))))))))(((......)))..........))))))........ ( -27.40) >DroEre_CAF1 32836 109 - 1 AUACAUACACGUCUACA-ACGCAUAUAAAUGGCCUCAGACAAAAGACUUUCCUUGCCAAGGACCAUUUGUACGCAGACAAAUUUCUAAAAAUACUUGCGGCUGACACUCU ..........(((....-.((((((((((((((((..(....(((......))).)..))).)))))))))...(((......))).........))))...)))..... ( -21.60) >DroYak_CAF1 24644 109 - 1 AUAUAAGCACGUUUACA-ACGUAUAUAAAUGGCCUCAGACAAUAGACUGUCCUUGCCAAGGACCAUUUGUGCGCAGACAAAUUUCUAAAAAUAAUUGCGGUUGACAUUCU ...............((-((...((((((((((((((((((......))))..))...))).)))))))))(((((....((((....))))..)))))))))....... ( -20.90) >consensus AUAUAAGCACGUUUACA_ACGUAUAUAAAUGGCCUCAGACAAUAGACUUUCCUUGUCAAGGACCAUUUGUACGCAGACAAAUUUCUAAAAAUACUUGCGGUUGACAUUCU .....(((.(((........((.((((((((((((..(((((..........))))).))).))))))))).))(((......)))..........))))))........ (-19.06 = -19.38 + 0.32)



| Location | 25,008,572 – 25,008,691 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -17.72 |
| Energy contribution | -18.76 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25008572 119 - 27905053 GGAAAACAAUGCACUACUUCAACCACAACCCUGAUCUCAUAUAUAAGCACGUUUACA-ACGUAUAUAAAUGGCCUCAGACAAUAGACUCUCCUUGUCAAGGACCAUUUGUAAGCAGACAA .........(((....................................((((.....-)))).((((((((((((..(((((..........))))).))).))))))))).)))..... ( -21.50) >DroSec_CAF1 31000 120 - 1 GGAAAACAAUGCACUACUUCAGCCACAACCCUGAUCUCAUAUAUAAGCACGUUUACAAACGUAUAUAAAUGGCCUCAGACAAUAGACUUUCCUUGUCAAGGACCAUUUGUAUGCAGACGA ..................((((........))))...............(((((......(((((((((((((((..(((((..........))))).))).))))))))))))))))). ( -25.30) >DroSim_CAF1 25114 120 - 1 GGAAAACAAUGCACUACUUCAGCCACAACCCUGAUCUCAUAUAUAAGCACGCUUACAAACGUAUAUAAAUGGCCUCAGACAAUAGACUUUCCUUGUCAAGGACCAUUUGUAUGCAGACAA ..................((((........))))(((......((((....)))).....(((((((((((((((..(((((..........))))).))).)))))))))))))))... ( -25.60) >DroEre_CAF1 32866 119 - 1 GCACAACAAUGCACUACUUCAACCACAACCCUGAUCUCAUAUACAUACACGUCUACA-ACGCAUAUAAAUGGCCUCAGACAAAAGACUUUCCUUGCCAAGGACCAUUUGUACGCAGACAA (((......)))......................................((((...-..((.((((((((((((..(....(((......))).)..))).))))))))).)))))).. ( -21.20) >DroYak_CAF1 24674 119 - 1 GCACAACAAUGCACUACUUCAGCCGCAACCCUGAUCCCAUAUAUAAGCACGUUUACA-ACGUAUAUAAAUGGCCUCAGACAAUAGACUGUCCUUGCCAAGGACCAUUUGUGCGCAGACAA (((......))).........(((((((..((((..((((.((((...((((.....-)))).)))).))))..))))..........(((((.....)))))...))))).))...... ( -27.00) >consensus GGAAAACAAUGCACUACUUCAGCCACAACCCUGAUCUCAUAUAUAAGCACGUUUACA_ACGUAUAUAAAUGGCCUCAGACAAUAGACUUUCCUUGUCAAGGACCAUUUGUACGCAGACAA .........(((......((((........))))..............((((......)))).((((((((((((..(((((..........))))).))).))))))))).)))..... (-17.72 = -18.76 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:44 2006