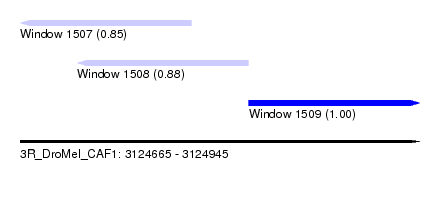
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,124,665 – 3,124,945 |
| Length | 280 |
| Max. P | 0.999317 |

| Location | 3,124,665 – 3,124,785 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.12 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3124665 120 - 27905053 CCAAACCAGCCCGGCAUCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGCAUGCUUCUUAAAGGAUAUUCGAAGUUAAUUAAUUUUUAAA ........(((((((......))))...(((((((....))))))).)))(((..((.(((((.....(((.......)))......)))))))...))).................... ( -26.90) >DroSec_CAF1 356 120 - 1 CCAAACCAGCCCGGCAGCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGCAUGCUUCUUAAAGGAUAUUCGAAGUUAAUUAAUUUUUAAA ........(((((((......))))...(((((((....))))))).)))(((..((.(((((.....(((.......)))......)))))))...))).................... ( -26.90) >DroSim_CAF1 726 120 - 1 CCAAACCAGCCCGGCAGCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGCAUGCUUCUUAAAGGAUAUUCCAAGUUAAUUAAUUUUUAAA ........(((((((......))))...(((((((....))))))).))).............((((((((.......)))...........((.....))))))).............. ( -26.70) >DroEre_CAF1 455 119 - 1 CCA-CCCAGCCCGGCAUCCCCGCUGAAUGGACUCGUUAGCGAGUUCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGCAUGGUUCUUAAAGGAUAUUCGAAGUUAAUUAAUUUUUAAA ...-....(((((((......))))...(((((((....))))))).)))(((..((.(((((((((.(((.......))).)))).)))))))...))).................... ( -28.40) >DroYak_CAF1 581 119 - 1 CCA-CCCUGCCCGGAAUCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAAUCCAUUUAAAACUUUGCUACACCCGCAUGGUUCUUAAAGGAUAUUCGAAGUUAAUUAAUUUUUAAA ...-....((((((.....)))......(((((((....))))))).)))(((((((.(((((((((.(((.......))).)))).))))))))).))).................... ( -32.60) >consensus CCAAACCAGCCCGGCAUCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGCAUGCUUCUUAAAGGAUAUUCGAAGUUAAUUAAUUUUUAAA ........(((((((......))))...(((((((....))))))).)))(((..((.(((((.....(((.......)))......)))))))...))).................... (-25.08 = -25.12 + 0.04)

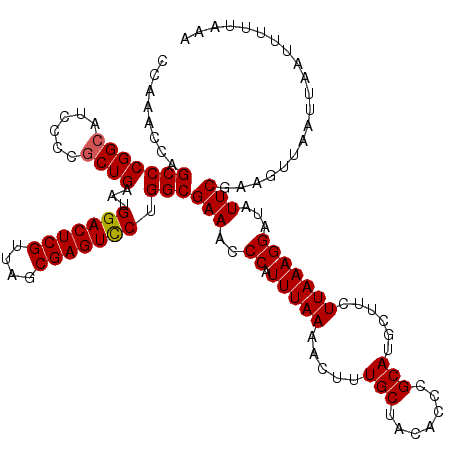
| Location | 3,124,705 – 3,124,825 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -32.82 |
| Energy contribution | -32.86 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3124705 120 - 27905053 CGACGUGUCCAGGCAGAAAGGUUCACCUUGGCUCUCCACCCCAAACCAGCCCGGCAUCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGC ....((((...((((((..((((.....(((....))).....)))).(((((((......))))...(((((((....))))))).)))................)))))))))).... ( -35.20) >DroSec_CAF1 396 120 - 1 CGACGUGUCCAGGCAGAAAGGUUCACCUUGGCUCUCCACCCCAAACCAGCCCGGCAGCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGC ....((((...((((((..((((.....(((....))).....)))).(((((((......))))...(((((((....))))))).)))................)))))))))).... ( -35.20) >DroSim_CAF1 766 120 - 1 CGACGUGUCCAGGCAGAAAGGUUCACCUUGGCUCUCCACCCCAAACCAGCCCGGCAGCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGC ....((((...((((((..((((.....(((....))).....)))).(((((((......))))...(((((((....))))))).)))................)))))))))).... ( -35.20) >DroEre_CAF1 495 119 - 1 CGACGUGUCCAGGCAGAAAGGUUCACCUUGGAUCGCCACCCCA-CCCAGCCCGGCAUCCCCGCUGAAUGGACUCGUUAGCGAGUUCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGC ....((((...((((((((((....))))((.((((((.....-..((((..((....)).))))...(((((((....)))))))))))))...)).........)))))))))).... ( -37.10) >DroYak_CAF1 621 119 - 1 CGACGUGUCCAGGCAGAAAGGUUCACCUUGGAUCUCCACCCCA-CCCUGCCCGGAAUCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAAUCCAUUUAAAACUUUGCUACACCCGC ....((((((.(((((.((((....))))((....))......-..))))).))).....((((....(((((((....))))))).))))......................))).... ( -37.40) >consensus CGACGUGUCCAGGCAGAAAGGUUCACCUUGGCUCUCCACCCCAAACCAGCCCGGCAUCCCCGCUGAAUGGACUCGUUAGCGAGUCCUGGCGAAACCCAUUUAAAACUUUGCUACACCCGC ....((((...((((((((((....))))((.((.(((........((((..((....)).))))...(((((((....)))))))))).))...)).........)))))))))).... (-32.82 = -32.86 + 0.04)

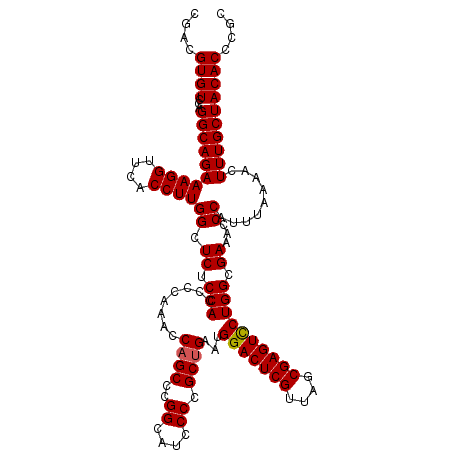
| Location | 3,124,825 – 3,124,945 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.95 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -24.56 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3124825 120 + 27905053 UCGUCCUUGGAGACAAUCACUCUCCAUUCAAAACAAUAAUUUCGCUGCUCCUUGAAAAAAAGCGUUGGGAAACUCGCUGGAGCUCAAUGAAAAGCAAAAAGCAAGAGGAAAACAAAGAAA ...((((((((((.......)))))..............(((((..(((((((....)).((((..((....)))))))))))....))))).((.....))..)))))........... ( -29.10) >DroSec_CAF1 516 119 + 1 UCGUCCUUGGAGACAAUCACUCUCCAUUCAAAACAAUAAUUUCGCUGCUCCUUGAAA-AAAGCGUUGGGAAACUCGCUGGAGCUCAAUGAAAAGCAAAAAGCAAGAGGAAAACAAAGAAA ...((((((((((.......)))))..............(((((..(((((((....-))((((..((....)))))))))))....))))).((.....))..)))))........... ( -29.10) >DroSim_CAF1 886 120 + 1 UCGUCCUUGGAGACAAUCACUCUCCAUUCAAAACAAUAAUUUCGCUGCUCCUUGAAAAAAAGCGUUGGGAAACUCGCUGGAGCUCAAUGAAAAGCAAAAAGCAAGAGGAAAACAAAGAAA ...((((((((((.......)))))..............(((((..(((((((....)).((((..((....)))))))))))....))))).((.....))..)))))........... ( -29.10) >DroEre_CAF1 614 112 + 1 UCGUCCUUGGAGACAAUCACUCUCCAUUCAAAACAAUAAUUUCGCUGCUCCUUGAAA-AAACUGCUGGGAAACUCGCUGGAGCUCAAUGAAAG-------GCAGGAGGAAAACAAAGAAA ((.((((((((((.......)))))).............(((((..(((((((....-))...((.((....)).)).)))))....))))).-------..)))).))........... ( -28.70) >DroYak_CAF1 740 112 + 1 UCGUCCUUGGAGACAAUCACUCUCCAUUCAAAACAAUAAUUUCGCUGCUCCUUGAAA-AAACUGUUGGGAAACUCGCUGGAGCUCAAUGAAAA-------GCAAGAGGAAAACAAAGAAA ...((((((((((.......)))))..............(((((..(((((((....-))...((.((....)).)).)))))....))))).-------....)))))........... ( -25.20) >consensus UCGUCCUUGGAGACAAUCACUCUCCAUUCAAAACAAUAAUUUCGCUGCUCCUUGAAA_AAAGCGUUGGGAAACUCGCUGGAGCUCAAUGAAAAGCAAAAAGCAAGAGGAAAACAAAGAAA ...((((((((((.......)))))..............(((((..(((((.........((((..((....)))))))))))....)))))............)))))........... (-24.56 = -24.72 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:13 2006