| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,003,075 – 25,003,166 |
| Length | 91 |
| Max. P | 0.754254 |

| Location | 25,003,075 – 25,003,166 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.58 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25003075 91 - 27905053 GUAGGAAUGGCAUUGAUGUUUGUUAUUAUUUGUGUCGUCCUGCGAUUGUUUAGCCAGUGAGUAAAUUGCAAUGCAAACAUCUCCAU------CUCCA------------------- ((((((.((((((.((((........)))).))))))))))))(((.((((.((..(..(.....)..)...))))))))).....------.....------------------- ( -22.80) >DroVir_CAF1 44386 108 - 1 GUAGGAAUGGCAUUGAUGUUUAUUAUUAUUUGUGUCGUCCUGCGAUUGUUUAGCCAGUGAGUAAAUUGCAAUGCAAACAUUU--AU------CACUUAUCACUUAUCACCCUAAGC ((((((.((((((.((((........)))).))))))))))))(((.........(((((.((((((((...)))...))))--))------))))........)))......... ( -23.83) >DroGri_CAF1 28453 90 - 1 GUAGGAAUGGCAUUGAUGUUUAUUAUUAUUUGUGUCGUCCUGCGAUUGUUUAGCCAGUGAGUAAAUUGCAAUGCAAAUAUUU--AU------CACCAA------------------ ((((((.((((((.((((........)))).)))))))))))).............((((.((((((((...)))...))))--))------)))...------------------ ( -21.50) >DroSec_CAF1 25626 97 - 1 GUAGGAAUGGCAUUGAUGUUUGUUAUUAUUUGUGUCGUCCUGCGAUUGUUUAGCCAGUGAGUAAAUUGCAAUGCAAACAUCUCCAUCUCCAUCUCCA------------------- ((((((.((((((.((((........)))).))))))))))))(((.((((.((..(..(.....)..)...)))))))))................------------------- ( -22.80) >DroWil_CAF1 29621 89 - 1 GUAGGAAUGGCAUUGAUGUUUAUUAUUAUUUGUGUCGUCCUGCGAUUGUUUAGCCAGUGAGUAAAUUGCAAAGCAAACAUUCCAUU------CAC--------------------- ((((((.((((((.((((........)))).))))))))))))((.(((((.((..(..(.....)..)...))))))).))....------...--------------------- ( -23.90) >DroYak_CAF1 19058 94 - 1 GUAGGAAUGGCAUUGAUGUUUGUUAUUAUUUGUGUCGUCCUGCGAUUGUUUAGCCAGUGAGUAAAUUGCAAUGCAAACAUCUCCAUCCCCCG---CA------------------- ((((((.((((((.((((........)))).))))))))))))(((.((((.((..(..(.....)..)...)))))))))...........---..------------------- ( -22.80) >consensus GUAGGAAUGGCAUUGAUGUUUAUUAUUAUUUGUGUCGUCCUGCGAUUGUUUAGCCAGUGAGUAAAUUGCAAUGCAAACAUCUCCAU______CACCA___________________ ((((((.((((((.((((........)))).))))))))))))...(((((.((..(..(.....)..)...)))))))..................................... (-21.72 = -21.58 + -0.14)

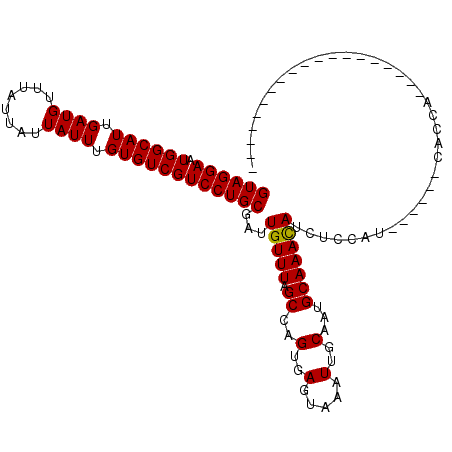

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:39 2006