| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,001,064 – 25,001,157 |
| Length | 93 |
| Max. P | 0.739555 |

| Location | 25,001,064 – 25,001,157 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.43 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.03 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
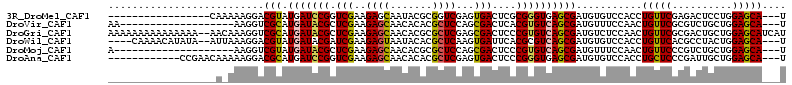
>3R_DroMel_CAF1 25001064 93 - 27905053 -----------------CAAAAAGGACGUAUGAUCCGGUCGAAGAGCAAUACGCGGUCGAGUGACUCGCGGGUGAGCGAUGUGUCCACCUGUUCGAGACUCCUGGAGCA---U -----------------............(((.(((((((...(((((...(((((((....))).))))((((..(.....)..)))))))))..))))...))).))---) ( -29.70) >DroVir_CAF1 24793 91 - 1 AA-------------------AAGGUCGCAUGAUACGCUCGAAGAGCAACACACGCUCCAGCGACUCACGUGUCAGCGAUGUUUCCAACUGUUCGCGUCUGCUGGAGCA---U ..-------------------...............((((...)))).......(((((((((((......))).((((.((.....))...))))....)))))))).---. ( -30.30) >DroGri_CAF1 25768 111 - 1 AAAAAAAAAAAAAAA--AACAAAGGUCGCAUGAUACGCUCGAAGAGCAACACGCGCUCGAGCGACUCCCGUGUCAGCGAUGUCUCCAACUGUUCGCGACUGCUGGAGCAUCAU ...............--..........((.((((((((((((.(.((.....)).)))))))(.....)))))))))((((.(((((.(.(((...))).).))))))))).. ( -33.40) >DroWil_CAF1 26914 104 - 1 ----CAAAACAUAUA--AUUAAAGGACGUAUGAUACGAUCGAAGAGUAAUACACGCUCAAGUGAUUCACGCGUCAGCGAUGUGUCCACCUGUUCACGCCUACUGGAGCA---U ----...........--......((((((((.....(((((..((((.......))))...)))))..(((....)))))))))))...(((((.((.....)))))))---. ( -20.90) >DroMoj_CAF1 76208 90 - 1 A--------------------AAGGUCGUAUGAUACGCUCGAAGAGCAACACGCGCUCCAGCGACUCCCGUGUCAGCGAUGUUUCCAACUGUUCCCGUCUGCUGGAGCA---U (--------------------((.(((((.((((((((((...))))....(((......)))......))))))))))).))).....((((((.(....).))))))---. ( -29.50) >DroAna_CAF1 28587 98 - 1 ------------CCGAACAAAAAGGACGCAUGAUCCGGUCGAAGAGCAACACACGCUCGAGUGACUCCCGGGUGAGCGAUGUGUCCACCUGCUCCCGAUUGCUGGAGCA---U ------------...........((((((((.(((((((((..((((.......))))...)))...)))))).)....)))))))...((((((.(....).))))))---. ( -35.10) >consensus __________________A_AAAGGACGCAUGAUACGCUCGAAGAGCAACACACGCUCGAGCGACUCCCGUGUCAGCGAUGUGUCCAACUGUUCGCGACUGCUGGAGCA___U ..........................(((.(((((((.(((..((((.......))))...)))....))))))))))...........(((((..........))))).... (-19.48 = -19.03 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:38 2006