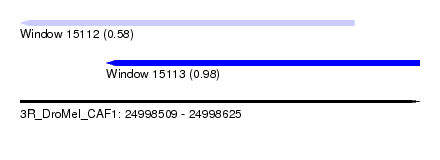
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,998,509 – 24,998,625 |
| Length | 116 |
| Max. P | 0.980815 |

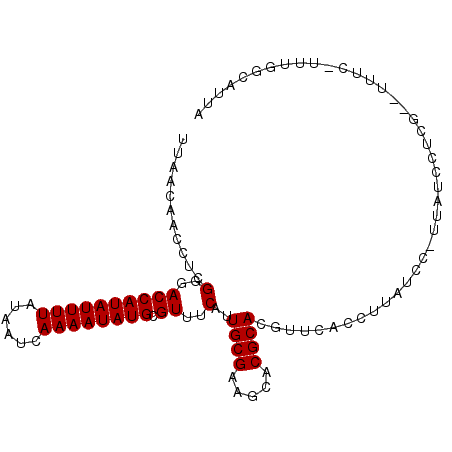
| Location | 24,998,509 – 24,998,606 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -15.60 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24998509 97 - 27905053 UUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAUUGCGAAGCACGCACGUUCACCUUAUAC-UUAUCCUCG--UUUCUUUUGGCAUUA .......((..(..((((((((((......)))))))).))..)..((((.....))))..............-.........--........))..... ( -14.80) >DroPse_CAF1 21005 92 - 1 UAAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAUUGCGAAGCACGCACGUUCAGAUUAUCC-UUGUUUUCAAAUUUCAUUU------- .((((((....(..((((((((((......)))))))).))..)..((((.....))))..............-)))))).............------- ( -16.30) >DroSim_CAF1 14355 97 - 1 UUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAUUGCGAAGCACGCACGUUCACCUUAUCC-UUAUCCUCG--UUUCCUUCGGCAUUA ...........(..((((((((((......)))))))).))..)..(((((((.....(((............-.......))--)...)))).)))... ( -15.11) >DroEre_CAF1 22447 87 - 1 UUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAUUGCGAAGCACGCACGUACAC-------CUUAUCCUCG--UU----UCGGCAUUA ...........(..((((((((((......)))))))).))..)..((((((((............-------.........)--))----)).)))... ( -15.40) >DroAna_CAF1 25868 90 - 1 UUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAUUGCGAUGCACGCACGUUCACCA------UUAUUCUCG--UUU--UUGGAUUUUA ..(((......(..((((((((((......)))))))).))..)..((((.....)))).)))..(((------..(......--.).--.)))...... ( -15.70) >DroPer_CAF1 30714 92 - 1 UAAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAUUGCGAAGCACGCACGUUCAGAUUAUCC-UUGUUUUCAAAUUUCAUUU------- .((((((....(..((((((((((......)))))))).))..)..((((.....))))..............-)))))).............------- ( -16.30) >consensus UUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAUUGCGAAGCACGCACGUUCACCUUAUCC_UUAUCCUCG__UUUC_UUUGGCAUUA ...........(..((((((((((......)))))))).))..)..((((.....))))......................................... (-14.00 = -14.00 + 0.00)


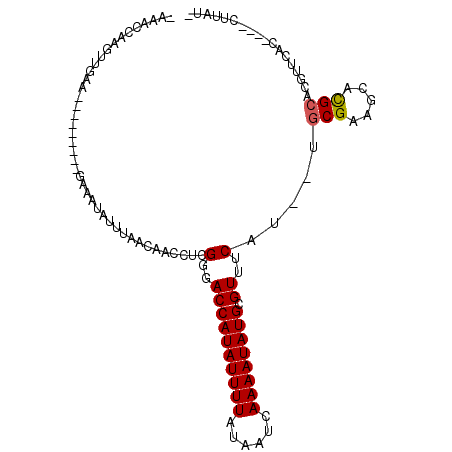
| Location | 24,998,534 – 24,998,625 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -17.07 |
| Consensus MFE | -12.09 |
| Energy contribution | -12.12 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24998534 91 - 27905053 --AACCAAGUUGAA---------GAAAUAUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAU--UGCGAAGCACGCACGUUCAC----CUUAUA --......((((((---------......))))))......(..((((((((((......)))))))).))..)..--((((.....)))).......----...... ( -17.90) >DroVir_CAF1 22183 95 - 1 CACACACAGAUAUA---------AAAAUAUUAAAAAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAAUUAACGAAGCAUGCACCUUUAG----UUCCCA ........(((((.---------...)))))..........((((.((((((((......)))))))).(((((.......)))))............----.)))). ( -14.70) >DroEre_CAF1 22468 85 - 1 --AACCAAGUUGAA---------GAAAUAUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAU--UGCGAAGCACGCACGUACAC---------- --......((((((---------......))))))......(..((((((((((......)))))))).))..)..--((((.....)))).......---------- ( -17.90) >DroWil_CAF1 24609 94 - 1 AAAACCAAAAAAA-----CAAAAAGAAAAUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAA--AGCGAAGCACGCAAGUUUU-------UAUU ........(((((-----(......................(..((((((((((......)))))))).))..)..--.(((.....)))..)))))-------)... ( -17.20) >DroYak_CAF1 14385 105 - 1 -CAACCAAGUUGAAGAAAUUGAAGAAAUAUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAU--UGCGAAGCACGCACGUACACUAACCUUAUC -.......((((((...(((.....))).))))))......(..((((((((((......)))))))).))..)..--((((.....))))................. ( -17.00) >DroAna_CAF1 25890 91 - 1 AAAACCAAGUUGAAGA-------GAGAUAUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAU--UGCGAUGCACGCACGUUCAC----CA---- ........((((((..-------......))))))......(..((((((((((......)))))))).))..)..--((((.....)))).......----..---- ( -17.70) >consensus _AAACCAAGUUGAA_________GAAAUAUUUAACAACCUCGGGACCAUAUUUUAUAAUCAAAAUAUGCGUUUCAU__UGCGAAGCACGCACGUUCAC____CUUAU_ .........................................(..((((((((((......)))))))).))..).....(((.....))).................. (-12.09 = -12.12 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:37 2006