
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,997,989 – 24,998,085 |
| Length | 96 |
| Max. P | 0.627555 |

| Location | 24,997,989 – 24,998,085 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.61 |
| Mean single sequence MFE | -15.63 |
| Consensus MFE | -11.03 |
| Energy contribution | -11.03 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24997989 96 - 27905053 ACAGACGAUUCUCAUUUGCUUUUGUGCUCAAUCAAAGAAGCUUAACUGUGAGAAUACCAAAAUGAAUACAAGAUACAAAUAAACGUAGAGCGAAA-------A------- .....((((((((((..((((((.((......)).))))))......))))))))..................(((........)))...))...-------.------- ( -13.90) >DroVir_CAF1 21536 108 - 1 ACAA--AACUCUCAUUUGCUUUUGUGCUCAAUCAAAGAAGCUUAACUGUGAGAAUAACAAAAUGAAUACAAAACGUGUUUGUACUUAAAACAAAAUGAAACAAAAUCAUU ....--...((((((..((((((.((......)).))))))......))))))........(..(((((.....)))))..)...........(((((.......))))) ( -16.10) >DroPse_CAF1 20414 99 - 1 ACAGAGAACUCUCAUUUGCUUUUGUGCUCAAUCAAAGAAGCUUAACUGUGAGAAUAACAAAACGAAUACAAGAUACAAAUAACUGUAGAGCAAGA-------AAAU---- ...(((....))).(((((((((((.((((.....((........)).))))....)))).............((((......))))))))))).-------....---- ( -15.40) >DroMoj_CAF1 72432 106 - 1 ACAAAAAACUCUCAUUUGCUUUUGUGCUCAAUCAAAGAAGCUUAACUGUGAGAAUAAGAAAGUGAAUACAAAAUGUGUUUAUACUUAAAAAAUAAACAUACAA----AUG .........((((((..((((((.((......)).))))))......)))))).((((...((((((((.....)))))))).))))................----... ( -17.90) >DroAna_CAF1 25292 97 - 1 ACCGACAAUUCUCAUUUGCUUUUGUGCUCAAUCAAAGAAGCUUAACUGUGAGAAUACCAAAAUGAAUACAAGAUACAAAUAAAUGUAGAGCGAAA-G-----A------- .......((((((((..((((((.((......)).))))))......))))))))..................((((......))))...(....-)-----.------- ( -15.10) >DroPer_CAF1 30124 99 - 1 ACAGAGAACUCUCAUUUGCUUUUGUGCUCAAUCAAAGAAGCUUAACUGUGAGAAUAACAAAACGAAUACAAGAUACAAAUAACUGUAGAGCAAGA-------AAAU---- ...(((....))).(((((((((((.((((.....((........)).))))....)))).............((((......))))))))))).-------....---- ( -15.40) >consensus ACAGACAACUCUCAUUUGCUUUUGUGCUCAAUCAAAGAAGCUUAACUGUGAGAAUAACAAAAUGAAUACAAGAUACAAAUAAACGUAGAGCAAAA_______A_______ .........((((((..((((((.((......)).))))))......))))))......................................................... (-11.03 = -11.03 + -0.00)

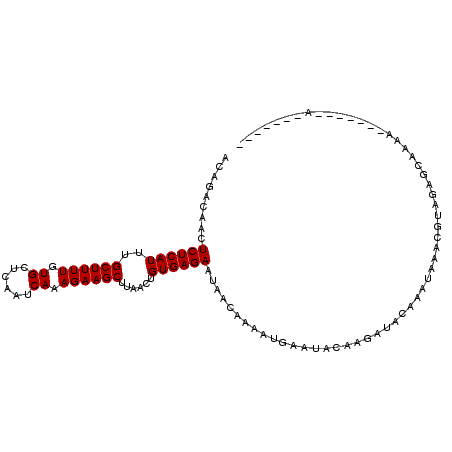
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:34 2006