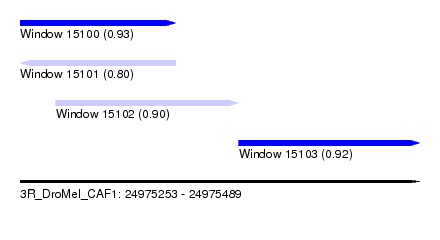
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,975,253 – 24,975,489 |
| Length | 236 |
| Max. P | 0.929671 |

| Location | 24,975,253 – 24,975,345 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -14.81 |
| Energy contribution | -14.53 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
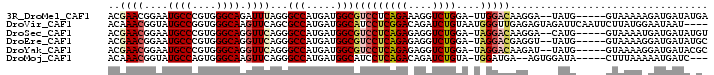
>3R_DroMel_CAF1 24975253 92 + 27905053 UCAUAUCAUCUUUUUAC-----CAUA--UCCUUGUCCAA-UCCAGACCUUUCUGAGGACGCCAUCAUGGCCCUAAAUCUGCCCACGGGCAUUCCGUUCGU .................-----....--.....((((..-..((((....)))).))))(((.....)))........((((....)))).......... ( -17.80) >DroVir_CAF1 1965 96 + 1 ----AUUAUUCCAUAAGAAUUGAAUCUACUCUCAACCCAUUACAGAUCUGUCCGAGGAUGCCAUCAUGGCGCUGAACUUGCCCACCGGCAUACCGUUUGU ----...............((((........))))......(((((((.....))(((((((....(((.((.......)))))..))))).)).))))) ( -15.40) >DroSec_CAF1 2350 92 + 1 ACAUAUCAUCAUUUUAC-----CAUG--UCCUUGUCCUA-UCCAGACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGU ................(-----((((--....((((((.-..((((....))))))))))....)))))....((((.((((....))))....)))).. ( -25.40) >DroEre_CAF1 2478 92 + 1 GCAUAUCAUCCUUUUAC-----CAUA--ACCUCGUCCUA-UCCAGACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGU ................(-----(((.--....((((((.-..((((....)))))))))).....))))....((((.((((....))))....)))).. ( -23.40) >DroYak_CAF1 2456 92 + 1 GCGUAUCAUCCUUUUAC-----CAUA--AUCUUGUCCUA-UCCAGACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGU ................(-----(((.--((..((((((.-..((((....))))))))))..)).))))....((((.((((....))))....)))).. ( -22.50) >DroMoj_CAF1 1965 89 + 1 ---GAUCAUUUUUAAAG-----UAUCCACU--UCAUCCA-UACAGAUCUGUCUGAGGAUGCCAUCAUGGCCCUGAACUUGCCCACUGGCAUACCGUUUGU ---..............-----........--.((..(.-..((((....)))).((((((((....(((.........)))...)))))).)))..)). ( -17.10) >consensus _CAUAUCAUCCUUUUAC_____CAUA__ACCUUGUCCUA_UCCAGACCUCUCUGAGGACGCCAUCAUGGCCCUGAACCUGCCCACGGGCAUUCCGUUCGU ..........................................((((....)))).(((((((.....)))........((((....))))....)))).. (-14.81 = -14.53 + -0.28)



| Location | 24,975,253 – 24,975,345 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -17.44 |
| Energy contribution | -18.17 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24975253 92 - 27905053 ACGAACGGAAUGCCCGUGGGCAGAUUUAGGGCCAUGAUGGCGUCCUCAGAAAGGUCUGGA-UUGGACAAGGA--UAUG-----GUAAAAAGAUGAUAUGA ...........((((((((.(........).)))))..)))(((((((((....))))).-..))))....(--(((.-----((......)).)))).. ( -21.80) >DroVir_CAF1 1965 96 - 1 ACAAACGGUAUGCCGGUGGGCAAGUUCAGCGCCAUGAUGGCAUCCUCGGACAGAUCUGUAAUGGGUUGAGAGUAGAUUCAAUUCUUAUGGAAUAAU---- ......((.((((((((((((.......)).))))..)))))))).........(((((((.((((((((......))))))))))))))).....---- ( -26.70) >DroSec_CAF1 2350 92 - 1 ACGAACGGAAUGCCCGUGGGCAGGUUCAGGGCCAUGAUGGCGUCCUCAGAGAGGUCUGGA-UAGGACAAGGA--CAUG-----GUAAAAUGAUGAUAUGU ..((((....((((....)))).))))...((((((.....(((((((((....))))..-.))))).....--))))-----))............... ( -27.40) >DroEre_CAF1 2478 92 - 1 ACGAACGGAAUGCCCGUGGGCAGGUUCAGGGCCAUGAUGGCGUCCUCAGAGAGGUCUGGA-UAGGACGAGGU--UAUG-----GUAAAAGGAUGAUAUGC ..((((....((((....)))).))))...((((((((..((((((((((....))))..-.))))))..))--))))-----))............... ( -33.80) >DroYak_CAF1 2456 92 - 1 ACGAACGGAAUGCCCGUGGGCAGGUUCAGGGCCAUGAUGGCGUCCUCAGAGAGGUCUGGA-UAGGACAAGAU--UAUG-----GUAAAAGGAUGAUACGC ..((((....((((....)))).))))...((((((((...(((((((((....))))..-.)))))...))--))))-----))............... ( -28.50) >DroMoj_CAF1 1965 89 - 1 ACAAACGGUAUGCCAGUGGGCAAGUUCAGGGCCAUGAUGGCAUCCUCAGACAGAUCUGUA-UGGAUGA--AGUGGAUA-----CUUUAAAAAUGAUC--- ......(((((.((.((((.(........).)))).((..(((((.((((....))))..-.))))).--.)))))))-----))............--- ( -21.80) >consensus ACGAACGGAAUGCCCGUGGGCAGGUUCAGGGCCAUGAUGGCGUCCUCAGAGAGGUCUGGA_UAGGACAAGAA__UAUG_____GUAAAAGGAUGAUAUG_ ..((((....((((....)))).))))...(((.....)))(((((((((....))))....)))))................................. (-17.44 = -18.17 + 0.72)

| Location | 24,975,274 – 24,975,382 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -31.31 |
| Consensus MFE | -23.01 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24975274 108 + 27905053 --------UCCUUGUCCAA-UCCAGACCUUUCUGAGGACGCCAUCAUGGCCCUAAAUCUGCCCACGGGCAUUCCGUUCGUCUACGAGCUGGACGAGAACUUCAAGCCGGUGGUGUCC --------...........-..((((....)))).((((((((((..(((........((((....)))).((((((((....))))).)))............))))))))))))) ( -38.20) >DroVir_CAF1 1987 111 + 1 ---UA---CUCUCAACCCAUUACAGAUCUGUCCGAGGAUGCCAUCAUGGCGCUGAACUUGCCCACCGGCAUACCGUUUGUAUACGAGCUGGAUGAGAACUUCAAGCCCGUUGUAUCC ---..---....((((.......((.((((((((.(((((((....(((.((.......)))))..))))).))(((((....)))))))))).))).))........))))..... ( -26.76) >DroGri_CAF1 697 114 + 1 ---UAUUGUUGUUAACCUCGCACAGAUCUCUCGGAGGAUGCCAUCAUGAAUCUGAACUUGCCAACUGGCAUUCCUUUUGUCUAUGAACUCGAUGAAAACUUCAAGCCAGUUGUCUCC ---................((((((((.((...((((...)).))..)))))))....)))((((((((.....(((..((.........))..))).......))))))))..... ( -21.80) >DroWil_CAF1 1917 116 + 1 GACAAUUGGCUUUUAACUU-UCUAGAUCUCUCUGAGGAUGCUAUUAUGGCUCUGAAUCUGCCCACUGGUAUCCCAUUCGUUUAUGAGUUGGAUGAAAACUUCAAGCCUGUGGUGUCC ((((...(((((.......-............((.((((((((....(((.........)))...))))))))))((((((((.....))))))))......))))).....)))). ( -29.40) >DroMoj_CAF1 1983 108 + 1 ---CA---CU--UCAUCCA-UACAGAUCUGUCUGAGGAUGCCAUCAUGGCCCUGAACUUGCCCACUGGCAUACCGUUUGUCUACGAGCUGGACGAGAACUUCAAGCCCGUGGUAUCC ---..---..--.......-..((((....)))).(((((((((...(((..((((..((((....)))).....((((((((.....))))))))...)))).))).))))))))) ( -32.60) >DroAna_CAF1 2309 98 + 1 ------------------U-UUUAGAUCUCUCUGAGGACGCCAUCAUGGCUCUGAACCUGCCCACGGGCAUCCCGUUCGUUUACGAGCUGGACGAGAACUUCAAGCCCGUGGUGUCG ------------------.-((((((....))))))((((((((...((((.((((..((((....))))(((.(((((....))))).))).......)))))))).)))))))). ( -39.10) >consensus ____A___CU_UUAACCCA_UACAGAUCUCUCUGAGGAUGCCAUCAUGGCUCUGAACCUGCCCACUGGCAUACCGUUCGUCUACGAGCUGGACGAGAACUUCAAGCCCGUGGUGUCC ......................((((....)))).(((((((((...(((..((((..((((....)))).....((((((((.....))))))))...)))).))).))))))))) (-23.01 = -23.18 + 0.17)


| Location | 24,975,382 – 24,975,489 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -28.57 |
| Energy contribution | -27.98 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
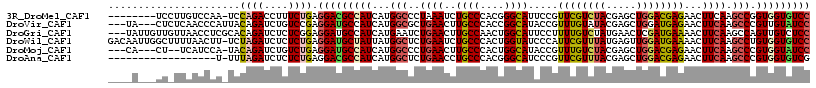
>3R_DroMel_CAF1 24975382 107 + 27905053 AUGCAGUUCCUCGGCGACGAGGAGACCGUGAAGAAGGCCAUCGAGGCUGUGGCCGCCCAGGGCAAGGCCAAGUAAGCA--CGCAGGCCAC------ACCUCAUG-AGAAUCC---ACUC ......(((((((....)))))))..(((((.(..((((..((..(((.((((((((...)))..)))))....))).--))..))))..------.).)))))-.......---.... ( -40.10) >DroGri_CAF1 811 87 + 1 AUGCAGUUCCUCGGCGACGAGGAGACCGUGAAGAAGGCCAUCGAAGCUGUCGCUGCCCAGGGCAAGGCCAAGUAAAUC--CA---UCUA-------AUC-------------------- .(((..(((((((....)))))))...........((((.....(((....)))(((...)))..))))..)))....--..---....-------...-------------------- ( -27.10) >DroEre_CAF1 2607 110 + 1 AUGCAGUUCCUCGGCGACGAGGAGACUGUGAAGAAGGCCAUCGAGGCUGUGGCUGCCCAGGGCAAGGCCAAGUAAGCA--GGCAGGCUAGCG----CCCUCAUGGAGAAUUC---ACCC ..(((((((((((....)))))).)))))...(((..((((.((((((.((((((((...)))..)))))....))).--(((........)----)))))))))....)))---.... ( -44.20) >DroYak_CAF1 2585 108 + 1 AUGCAGUUCCUCGGCGACGAGGAGACUGUGAAGAAGGCCAUCGAGGCGGUGGCUGCCCAGGGCAAGGCCAAGUAAGCA--CGCUGGCCAG------ACCGCAUGGAGAAUCC---ACUC ..(((((((((((....)))))).))))).............((((((((...((((...)))).(((((.((.....--.)))))))..------))))).(((.....))---)))) ( -43.70) >DroMoj_CAF1 2091 88 + 1 AUGCAGUUCCUUGGCGACGAGGAGACAGUCAAGAAGGCCAUCGAAGCUGUUGCUGCCCAGGGCAAGGCCAAGUAAACGCUAA-UCUCUA------------------------------ ..((..((.(((((((((..(....).)))......(((.....(((....)))......)))...)))))).))..))...-......------------------------------ ( -26.30) >DroAna_CAF1 2407 115 + 1 AUGCAGUUCCUCGGCGACGAGGAGACCGUGAAGAAGGCCAUUGAGGCUGUCGCCGCCCAGGGCAAGGCCAAGUAAGCU--AG-AGGCUAUCUACAUAUAUCAAU-GGAAUCCAACAUCC ......(((((((....)))))))...((...((...(((((((((((...(((......)))..))))..((((((.--..-..)))...))).....)))))-))..))..)).... ( -37.30) >consensus AUGCAGUUCCUCGGCGACGAGGAGACCGUGAAGAAGGCCAUCGAGGCUGUGGCUGCCCAGGGCAAGGCCAAGUAAGCA__CG_AGGCUA_______ACCUCAUG_AGAAUCC___AC_C .(((..(((((((....)))))))...........((((.....(((....)))(((...)))..))))..)))............................................. (-28.57 = -27.98 + -0.58)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:28 2006