| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,957,395 – 24,957,497 |
| Length | 102 |
| Max. P | 0.622448 |

| Location | 24,957,395 – 24,957,497 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -39.01 |
| Consensus MFE | -24.00 |
| Energy contribution | -23.87 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24957395 102 + 27905053 -CUCCAUUCGCAGA-UAUCGGGAAU--CCG--------------GCUGCCACAAUGGGUUCCCUAUUCCGCAGCGAGGAAAUGGCGCUGUGCCAGCUGUUCCUGCAGAGCGAGGCGGCCU -..((.(((((...-....((((((--(((--------------..........)))))))))...((.((((.(((.(..((((.....))))..).))))))).)))))))..))... ( -37.80) >DroPse_CAF1 19599 114 + 1 ----AAUUUGCAGAAAAUCGGUAAU--CAGGCGUUGCUUUUAAGGCUGCAAAGAUGGGUUCGUUGUUCCGUAGCGAGGAGAUGGCUUUGUGCCAGCUCUUCCUACAAAGUGAGGCUGCCU ----..(((((((......((((((--.....)))))).......)))))))...((((.(.(..((..((((.((((((.((((.....)))).))))))))))..))..).)..)))) ( -38.12) >DroSec_CAF1 18272 102 + 1 -CUCCAUUCGCAGA-UAUCGGGAAU--CCG--------------GCUGUCACAAUGGGUUCCCUGUUCCGCAGCGAGGAAAUGGCGCUGUGCCAGCUGUUCCUGCAGAGCGAGGCUGCCU -........((((.-..((((((((--(((--------------..........))))))))).((((.((((.(((.(..((((.....))))..).))))))).))))))..)))).. ( -41.30) >DroYak_CAF1 18336 103 + 1 CCUCCAUUCGCAGA-UAUCGGGAAU--CCG--------------GCUGCCACAAUGGGUUCCCUGUUCCGCAGCGAGGAAAUGGCUCUGUGCCAGCUGUUCCUGCAGAGCGAGGCGGCCU .....((((.(...-....).))))--..(--------------((((((.....((....))(((((.((((.(((.(..((((.....))))..).))))))).))))).))))))). ( -40.30) >DroAna_CAF1 19438 104 + 1 -AAAAAAUUCCAGA-CAUCAGUGACGACAG--------------GCGGCCACCAUGGGUUCCCUGUUCCGAAGCGAGGAAAUGGCACUGUGCCAGUUGUUCUUGCAGAGCGAGGCUGCCU -.............-.............((--------------((((((.....((....))(((((....(((((((..((((.....))))....))))))).))))).)))))))) ( -35.70) >DroPer_CAF1 19581 114 + 1 ----AAUUUGCAGAAAAUCGGUAAU--CAGGCGUUGCUUUUAAGGCUGCAAAGAUGGGUUCGUUGUUCCGUAGCGAGGAGAUGGCUUUGUGCCAGCUCUUCCUACAAAGCGAGGCUGCCU ----..(((((((......((((((--.....)))))).......)))))))...((((.(.(((((..((((.((((((.((((.....)))).))))))))))..))))).)..)))) ( -40.82) >consensus _CUCAAUUCGCAGA_UAUCGGGAAU__CAG______________GCUGCCACAAUGGGUUCCCUGUUCCGCAGCGAGGAAAUGGCUCUGUGCCAGCUGUUCCUGCAGAGCGAGGCUGCCU .........((((....(((........................(((((...(((((.....)))))..))))).(((((.((((.....))))....)))))......)))..)))).. (-24.00 = -23.87 + -0.14)

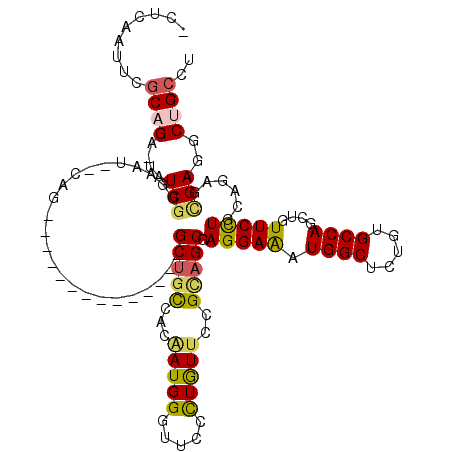

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:19 2006