
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,944,496 – 24,944,628 |
| Length | 132 |
| Max. P | 0.987070 |

| Location | 24,944,496 – 24,944,597 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 70.74 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
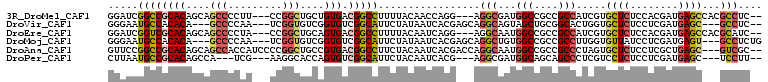
>3R_DroMel_CAF1 24944496 101 - 27905053 GGAUCGGCCGCACAGCAGCCCCUU---CCGGCUGCUGUGACGGCCUUUUACAACCAGG---AGGCGAUGGCCGCCGCCAUCGUGCUCUCCACGAUGAGCCACGCCUC-- .....(((((((((((((((....---..)))))))))).)))))............(---((((((((((....))))))((((((........))).))))))))-- ( -56.80) >DroVir_CAF1 5944 98 - 1 GGGAAUGCCACACA---GCCCCAA---UCGGUGUCGGUGUCGGCAUUCUAUAAUCACGAGCAGGCAGUAGCUGCGGCACUGGUGCUCUCCUCGAUGAGC---GCCUC-- .((((((((((((.---(((....---..)))....)))).))))))))..........((..((((...)))).))...(((((((........))))---)))..-- ( -39.30) >DroEre_CAF1 5575 101 - 1 GGAUCGGUCGCACAGCAGCCCCUA---CCGGCUGCAGUGACGGCCUUUUACAAUCAGG---AGGCAAUGGCCGCCGCCAUCGUGCUCUCCACGAUGAGCCACGCAUC-- ((.((((((((((.((((((....---..)))))).))).)))))...........((---((((((((((....)))))..))).)))).....)).)).......-- ( -43.30) >DroMoj_CAF1 5727 100 - 1 GGGAAUGCCACACA---GCCCCAA---UCGGUGUCGGUGUCGGCAUUCUAUAAUCACGAGCAGGCUGUGGCCGCCGCCUUGGUGUUAUCCUCGAUGAGU---GCCUCUG .((((((((((((.---(((....---..)))....)))).))))))))....(((((((..(((....)))..(((....))).....)))).)))..---....... ( -33.60) >DroAna_CAF1 5513 104 - 1 GUUCCGGCCGCACAGCAGCCACCAUCCCCGGCUGCCGUGACGGCCUUCUACAAUCACGACCAGGCAAUGGCCGCCGCCCUAGUGCUCUCCUCGCUGAGC---GUCGC-- .....((((((((.((((((.........)))))).))).)))))...........((((..(((....)))...........((((........))))---)))).-- ( -41.60) >DroPer_CAF1 5897 95 - 1 CUUAAUGCCGCACAGCCA---UCG---AAGGCACCAGUGUCGGCAUUCUACAAUCACG---AGGCGAUGGCAGCAGCCCUCGUCCUCUCCUCGAUGAGC---UCCUU-- ...((((((((((.(((.---...---..)))....))).)))))))......(((((---((((((.(((....))).)))......))))).)))..---.....-- ( -32.00) >consensus GGAAAGGCCGCACAGCAGCCCCAA___CCGGCUGCAGUGACGGCAUUCUACAAUCACG___AGGCAAUGGCCGCCGCCCUCGUGCUCUCCUCGAUGAGC___GCCUC__ .....((((((((....(((.........)))....))).))))).................(((...(((....))).....((((........))))...))).... (-19.24 = -19.72 + 0.47)


| Location | 24,944,534 – 24,944,628 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 71.35 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24944534 94 - 27905053 GCCCAGU---ACGCCCACGCGCCAAUGACAUUCGGGAUCGGCCGCACAGCAGCCCCUU---CCGGCUGCUGUGACGGCCUUUUACAACCAGG---AGGCGAUG .......---.((((..(...((..........))....(((((((((((((((....---..)))))))))).)))))...........).---.))))... ( -40.30) >DroVir_CAF1 5979 94 - 1 AGCUGGC---GCCUCCACGCAUCAAUGACAUACGGGGAAUGCCACACA---GCCCCAA---UCGGUGUCGGUGUCGGCAUUCUAUAAUCACGAGCAGGCAGUA .(((.((---((......))...............((((((((((((.---(((....---..)))....)))).))))))))..........)).))).... ( -28.10) >DroEre_CAF1 5613 94 - 1 GCCCAGU---ACGCCUUCGCGCCAAUGACAUUCGGGAUCGGUCGCACAGCAGCCCCUA---CCGGCUGCAGUGACGGCCUUUUACAAUCAGG---AGGCAAUG .......---..((((((...((..........))....((((((((.((((((....---..)))))).))).)))))...........))---)))).... ( -34.50) >DroYak_CAF1 5453 94 - 1 GCCAAAU---ACGCCUUCGCGCCAAUGACAUUCGGGAUCGGCCGCACAGCAGCCCCUA---CCGGCUGCAGUGACGGCCUUUUACAACCAGG---AGGCGAUG .......---.(((((((...((..........))....((((((((.((((((....---..)))))).))).)))))...........))---)))))... ( -39.20) >DroMoj_CAF1 5764 97 - 1 UGCCGGGAAACCAACAACGCAUCAAUGACAUACGGGGAAUGCCACACA---GCCCCAA---UCGGUGUCGGUGUCGGCAUUCUAUAAUCACGAGCAGGCUGUG .(((((....))......((.((..(((.......((((((((((((.---(((....---..)))....)))).))))))))....))).)))).))).... ( -33.00) >DroAna_CAF1 5548 100 - 1 GGCGGGC---ACCUCCCCGCGCCAAUGACAAACAGUUCCGGCCGCACAGCAGCCACCAUCCCCGGCUGCCGUGACGGCCUUCUACAAUCACGACCAGGCAAUG .(((((.---.....)))))(((...(((.....)))..((((((((.((((((.........)))))).))).))))).................))).... ( -41.40) >consensus GCCCAGU___ACGCCCACGCGCCAAUGACAUACGGGAUCGGCCGCACAGCAGCCCCUA___CCGGCUGCAGUGACGGCCUUCUACAAUCACG___AGGCAAUG ....................(((..((.....)).....((((((((.((((((.........)))))).))).))))).................))).... (-20.90 = -21.40 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:34:16 2006