
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,921,144 – 24,921,621 |
| Length | 477 |
| Max. P | 0.999689 |

| Location | 24,921,144 – 24,921,254 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.74 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -24.92 |
| Energy contribution | -26.36 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24921144 110 + 27905053 UUAUCCGCGAUUCCGCCAUUCAGUUGUGAAGCUGAGCACAUGGGCACACACUCGUGUG----------UGUGUGUGUGAGGUGCAUGGCGCUGUUGCUCUUGAUUUGGACUUGUGUGCCA ......(((((..((((((((((((....))))))((((..(.((((((((......)----------))))))).)...))))))))))..)))))........(((((....)).))) ( -44.00) >DroSec_CAF1 34264 104 + 1 UUAUCCGCGAUUCCGCCAUUCAGUUGUGAAGCUGAGCACAUGGGCACACACUCGUGUG----------------UGUGAGGUGCAUGGCGCUGUUGCUCUUGAUUUGGACUUGUGUGCCA ......(((((..((((((((((((....))))))((((.....(((((((....)))----------------))))..))))))))))..)))))........(((((....)).))) ( -41.30) >DroSim_CAF1 30001 104 + 1 UUAUCCGCGAUUCCGCCAUUCAGUUGUGAAGCUGAGCACAUGGGCACACACUCGUGUG----------------UGUGAGGUGCAUGGCGCUGUUGCUCUUGAUUUGGACUUGUGUGCCG ......(((((..((((((((((((....))))))((((.....(((((((....)))----------------))))..))))))))))..))))).........((((....)).)). ( -41.20) >DroEre_CAF1 33001 91 + 1 UUAUCCGCGAUUCCGCAAUUCAGUUGUGAAGCUGAGCACAUGUGCACACACUCGUA-----------------------------UGGCGCUGUUGCUCUUGAUUUGGACUUGUGUGCCA .....(((((.((((.((((.((((....))))(((((((.((((..((....)).-----------------------------..)))))).)))))..)))))))).)))))..... ( -23.90) >DroYak_CAF1 33413 120 + 1 UUAUCCGCGAUUCCCCCAUUCAGUUGUGAAGCUGAGCACAUGUGCACAUACUCGUAUGUACAUACAUAUGUAUGUGUGAGCUGCAUGGCGCUGUUGCUCUUGAUUUGGACUUGUGUGCCA .....(((((.(((.....((((((....))))))((.(((((((.(((((.(((((((....)))))))...))))).)).)))))))((....)).........))).)))))..... ( -37.60) >consensus UUAUCCGCGAUUCCGCCAUUCAGUUGUGAAGCUGAGCACAUGGGCACACACUCGUGUG________________UGUGAGGUGCAUGGCGCUGUUGCUCUUGAUUUGGACUUGUGUGCCA ......(((((..((((((((((((....)))))))((((((((......))))))))...........................)))))..)))))........(((((....)).))) (-24.92 = -26.36 + 1.44)



| Location | 24,921,214 – 24,921,334 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.41 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -36.18 |
| Energy contribution | -36.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24921214 120 - 27905053 GUGUGCUCUGCUCGUUGGAUGCUUGUUGCCUGAACGAUGUUUGUUUGCAGCCACCAAAGGAUUGGCAUCGGGAUUGGAUAUGGCACACAAGUCCAAAUCAAGAGCAACAGCGCCAUGCAC (.(((((.(((((.((((..((((((((((((..((((.((((..(((..((......))....))).))))))))..)).)))).)))))))))).....)))))..))))))...... ( -38.10) >DroSec_CAF1 34328 120 - 1 GUGUGCUCUGCUCGUUGGAUGCUUGUUGCCUGGACGAUGUUUGUUUGCCGCCACCAAAGGAUUGGCAUCGGGAUUGCAUAUGGCACACAAGUCCAAAUCAAGAGCAACAGCGCCAUGCAC (.(((((.(((((.((((..((((((((((((..((((.((((..((((.((......))...)))).))))))))..)).)))).)))))))))).....)))))..))))))...... ( -40.10) >DroSim_CAF1 30065 120 - 1 GUGUGCUCUGCUCGUUGGAUGCUUGUUGCCUGGACGAUGUUUGUUUGCAGCCACCAAAGGAUUGGCAUCGGGAUUGGAUACGGCACACAAGUCCAAAUCAAGAGCAACAGCGCCAUGCAC (.(((((.(((((.((((..((((((((((((..((((.((((..(((..((......))....))).))))))))..)).)))).)))))))))).....)))))..))))))...... ( -39.30) >DroEre_CAF1 33057 113 - 1 GUGUGC--UGCUCGUUGGAUGCUUGUUGCCUGGACGAUGUUUGCUUGCAGCCACCAAAGGAUUGGCAUCGGCAUUGGAUAUGGCACACAAGUCCAAAUCAAGAGCAACAGCGCCA----- (.((((--(((((.((((..((((((((((((..(((((((((((.....((......))...))))..)))))))..)).)))).)))))))))).....))))...)))))).----- ( -41.30) >DroYak_CAF1 33493 118 - 1 GUGUGC--UGCUCGUUGAAUGCUUGUUGCCUGGACGAUGUUUGUUUGCAGCCACCAAAGGAUUGGCAUCGAGAUUGGAUAUGGCACACAAGUCCAAAUCAAGAGCAACAGCGCCAUGCAG (.((((--(((((.((((..((((((((((((..((((.((((..(((..((......))....))).))))))))..)).)))).)))))).....))))))))...))))))...... ( -36.20) >consensus GUGUGCUCUGCUCGUUGGAUGCUUGUUGCCUGGACGAUGUUUGUUUGCAGCCACCAAAGGAUUGGCAUCGGGAUUGGAUAUGGCACACAAGUCCAAAUCAAGAGCAACAGCGCCAUGCAC (.((((..(((((.((((..((((((((((((..((((.((((..(((..((......))....))).))))))))..)).)))).)))))))))).....)))))...)))))...... (-36.18 = -36.42 + 0.24)



| Location | 24,921,254 – 24,921,374 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.66 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -37.06 |
| Energy contribution | -36.78 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24921254 120 - 27905053 UCCUGGCAACGAGUAAUGCGAGUCUGCCGCCUUUUGUUGUGUGUGCUCUGCUCGUUGGAUGCUUGUUGCCUGAACGAUGUUUGUUUGCAGCCACCAAAGGAUUGGCAUCGGGAUUGGAUA (((.(((((((((((..((((((..((((((.......).))).))...))))))....)))))))))))....((((((..((((............))))..)))))))))....... ( -40.80) >DroSec_CAF1 34368 120 - 1 UCCUGGCAACGAGUAAUGCGAGUCUGCUGCCUUUUGUUGUGUGUGCUCUGCUCGUUGGAUGCUUGUUGCCUGGACGAUGUUUGUUUGCCGCCACCAAAGGAUUGGCAUCGGGAUUGCAUA (((.(((((((((((..((((((..((((((.......).))).))...))))))....))))))))))).)))((((.((((..((((.((......))...)))).)))))))).... ( -40.10) >DroSim_CAF1 30105 120 - 1 UCCUGGCAACGAGUAAUGCGAGUCUGCUGCCUUUUGUUGUGUGUGCUCUGCUCGUUGGAUGCUUGUUGCCUGGACGAUGUUUGUUUGCAGCCACCAAAGGAUUGGCAUCGGGAUUGGAUA (((.(((((((((((..((((((..((((((.......).))).))...))))))....))))))))))).)))((((((..((((............))))..)))))).......... ( -39.60) >DroEre_CAF1 33092 118 - 1 UCCUGGCAACGAGUAAUGCGAGUCUGCUGCCUUUUGUUGUGUGUGC--UGCUCGUUGGAUGCUUGUUGCCUGGACGAUGUUUGCUUGCAGCCACCAAAGGAUUGGCAUCGGCAUUGGAUA (((.(((((((((((..((((((..((((((.......).))).))--.))))))....))))))))))).)))(((((((((((.....((......))...))))..))))))).... ( -42.80) >DroYak_CAF1 33533 118 - 1 UCCUGGCAACGGGUAAUGCGAGUCUGCUGUCUUUUGUUGUGUGUGC--UGCUCGUUGAAUGCUUGUUGCCUGGACGAUGUUUGUUUGCAGCCACCAAAGGAUUGGCAUCGAGAUUGGAUA (((.(((((((((((..((((((..((...(.........)...))--.))))))....))))))))))).)))((((((..((((............))))..)))))).......... ( -37.70) >consensus UCCUGGCAACGAGUAAUGCGAGUCUGCUGCCUUUUGUUGUGUGUGCUCUGCUCGUUGGAUGCUUGUUGCCUGGACGAUGUUUGUUUGCAGCCACCAAAGGAUUGGCAUCGGGAUUGGAUA (((.(((((((((((..((((((..((((((.......).))).))...))))))....))))))))))).)))((((((......)).((((((...))..)))))))).......... (-37.06 = -36.78 + -0.28)

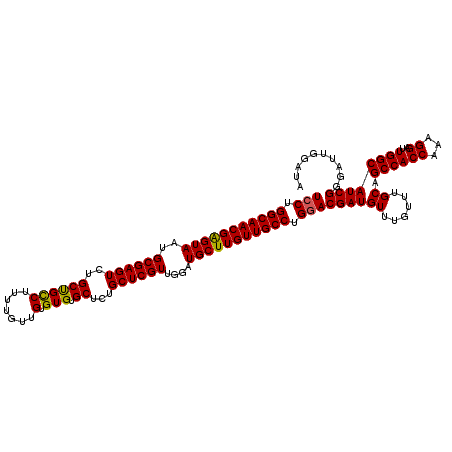

| Location | 24,921,334 – 24,921,454 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -41.24 |
| Consensus MFE | -34.76 |
| Energy contribution | -35.80 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24921334 120 + 27905053 ACAACAAAAGGCGGCAGACUCGCAUUACUCGUUGCCAGGAAUAUAUCACGGCCAACACAAACCCUCCCCUGGCCUUCUGGCCAUCUUGUGAUGAGGAUUGAAUGGCAACGACUGCUGCCA .........((((((((...........(((((((((..(((.((((((((..........))......(((((....)))))....))))))...)))...))))))))))))))))). ( -43.00) >DroSec_CAF1 34448 120 + 1 ACAACAAAAGGCAGCAGACUCGCAUUACUCGUUGCCAGGAAUAUAUCACGGCCAACACAAACCCUCCCCCGGCCUUCUGGCCAUCUUGUGAUGAGGAUUGAAUGGCAACGUCUGCUGCCA .........((((((((((..(.....)..(((((((((............))....(((..((((....((((....))))(((....))))))).)))..))))))))))))))))). ( -45.10) >DroSim_CAF1 30185 120 + 1 ACAACAAAAGGCAGCAGACUCGCAUUACUCGUUGCCAGGAAUAUAUCACGGCCAACACAAACCCUCCCCUGGCCUUCUGGCCAUCUUGUGAUGAGGAUUGAAUGGCAACGACUGCUGCCA .........((((((((...........(((((((((..(((.((((((((..........))......(((((....)))))....))))))...)))...))))))))))))))))). ( -43.70) >DroEre_CAF1 33170 119 + 1 ACAACAAAAGGCAGCAGACUCGCAUUACUCGUUGCCAGGAAUAUAUCACGGCCAACACAAUCCCUCCCCG-GGCUUCUGGCCAUCUUGUGAUGAGGAUUGAAUGGCAACGACUGCUGCCA .........((((((((...........(((((((((..(((.(((((((((((.......(((.....)-))....))))).....))))))...)))...))))))))))))))))). ( -44.60) >DroYak_CAF1 33611 110 + 1 ACAACAAAAGACAGCAGACUCGCAUUACCCGUUGCCAGGAAUAUAUCACGGCCAACUCAAUCCCCCCUC----------CCCAUCUUGUGAUGAGGAUUGAAUGGCAACGACUGCUGCCA .........(.((((((.((.(((........))).)).......((...((((..(((((((......----------..((((....)))).))))))).))))...)))))))).). ( -29.80) >consensus ACAACAAAAGGCAGCAGACUCGCAUUACUCGUUGCCAGGAAUAUAUCACGGCCAACACAAACCCUCCCCUGGCCUUCUGGCCAUCUUGUGAUGAGGAUUGAAUGGCAACGACUGCUGCCA .........((((((((...........(((((((((..(((.(((((((((((.......................))))).....))))))...)))...))))))))))))))))). (-34.76 = -35.80 + 1.04)



| Location | 24,921,334 – 24,921,454 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -48.20 |
| Consensus MFE | -40.87 |
| Energy contribution | -41.92 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24921334 120 - 27905053 UGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGGCCAGAAGGCCAGGGGAGGGUUUGUGUUGGCCGUGAUAUAUUCCUGGCAACGAGUAAUGCGAGUCUGCCGCCUUUUGUUGU .(((.((((((((((((......((((((((....)))((((....)))).)))))(((..((((((......))))))..)))))))))))(.....)....)))).)))......... ( -46.00) >DroSec_CAF1 34448 120 - 1 UGGCAGCAGACGUUGCCAUUCAAUCCUCAUCACAAGAUGGCCAGAAGGCCGGGGGAGGGUUUGUGUUGGCCGUGAUAUAUUCCUGGCAACGAGUAAUGCGAGUCUGCUGCCUUUUGUUGU .((((((((((((((((......((((((((....)))((((....)))).)))))(((..((((((......))))))..)))))))))..(.....)..))))))))))......... ( -52.90) >DroSim_CAF1 30185 120 - 1 UGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGGCCAGAAGGCCAGGGGAGGGUUUGUGUUGGCCGUGAUAUAUUCCUGGCAACGAGUAAUGCGAGUCUGCUGCCUUUUGUUGU .((((((((((((((((......((((((((....)))((((....)))).)))))(((..((((((......))))))..)))))))))))(.....)....))))))))......... ( -50.60) >DroEre_CAF1 33170 119 - 1 UGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGGCCAGAAGCC-CGGGGAGGGAUUGUGUUGGCCGUGAUAUAUUCCUGGCAACGAGUAAUGCGAGUCUGCUGCCUUUUGUUGU .((((((((((((((((......((((((((....)))(((.....)))-.)))))((((.((((((......)))))).))))))))))))(.....)....))))))))......... ( -47.20) >DroYak_CAF1 33611 110 - 1 UGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGGG----------GAGGGGGGAUUGAGUUGGCCGUGAUAUAUUCCUGGCAACGGGUAAUGCGAGUCUGCUGUCUUUUGUUGU .(((((((((((..((((((((((((((.((.(.....)..----------))..)))))))))).))))..)))..((((((((....)))).)))).....))))))))......... ( -44.30) >consensus UGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGGCCAGAAGGCCAGGGGAGGGUUUGUGUUGGCCGUGAUAUAUUCCUGGCAACGAGUAAUGCGAGUCUGCUGCCUUUUGUUGU .((((((((((((((((......((((((((....)))((((....)))).)))))(((..((((((......))))))..)))))))))))(.....)....))))))))......... (-40.87 = -41.92 + 1.05)



| Location | 24,921,374 – 24,921,488 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.35 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -31.58 |
| Energy contribution | -32.45 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24921374 114 - 27905053 UGUUUCUGUUCCUUGCCUUUUGAAUGCUCCCCGCUGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGGCCAGAAGGCCAGGGGAGGGUUUGUGUUGGCCGUGAUAUAU ......(((..(..(((...(((((.((((((..(((((((....)))))))...................(((((....))))))))))).)))))....))).)..)))... ( -40.70) >DroSec_CAF1 34488 114 - 1 UGUUUCUGCUCCUUGCCUUUUGAAUGCUCCCCGCUGGCAGCAGACGUUGCCAUUCAAUCCUCAUCACAAGAUGGCCAGAAGGCCGGGGGAGGGUUUGUGUUGGCCGUGAUAUAU ..............(((...(((((.((((((..(((((((....)))))))...................(((((....))))))))))).)))))....))).......... ( -39.30) >DroSim_CAF1 30225 114 - 1 UGUUACUGCUCCUUGCCUUUUGAAUGCUCCCCGCUGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGGCCAGAAGGCCAGGGGAGGGUUUGUGUUGGCCGUGAUAUAU ((((((.(((....((....(((((.((((((..(((((((....)))))))...................(((((....))))))))))).))))).)).))).))))))... ( -42.00) >DroYak_CAF1 33651 104 - 1 UCUUUCUGUUCCUUGCUUUUUGAAUGCUUCCCGCUGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGGG----------GAGGGGGGAUUGAGUUGGCCGUGAUAUAU ......(((..(..(((..((.(((.((((((..(((((((....))))))).....((((((((....))))))----------))))))))))).))..))).)..)))... ( -37.10) >consensus UGUUUCUGCUCCUUGCCUUUUGAAUGCUCCCCGCUGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGGCCAGAAGGCCAGGGGAGGGUUUGUGUUGGCCGUGAUAUAU ..............(((...(((((.((((((..(((((((....)))))))...................(((((....))))))))))).)))))....))).......... (-31.58 = -32.45 + 0.87)

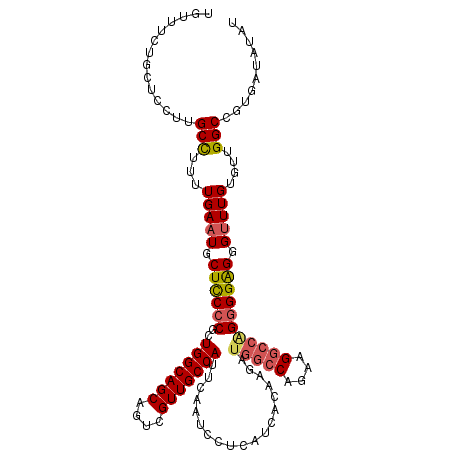

| Location | 24,921,414 – 24,921,520 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.71 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -25.75 |
| Energy contribution | -25.12 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24921414 106 - 27905053 GAGCGGAGGGCACCUCCUCCUCGUUCUCCUC---CUGUUUCUGUUCCUUGCCUUUUGAAUGCUCCCCGCUGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGG ((((((((((....))))))..)))).....---....................((((((((.....)).((((((....))))))))))))....((((....)))). ( -27.50) >DroSec_CAF1 34528 100 - 1 GAGCGGAGGGCACCUCCUCC------UCCUC---CUGUUUCUGCUCCUUGCCUUUUGAAUGCUCCCCGCUGGCAGCAGACGUUGCCAUUCAAUCCUCAUCACAAGAUGG ((((((((((....))))))------..(..---..).....))))........((((((((.....)).((((((....))))))))))))....((((....)))). ( -28.20) >DroSim_CAF1 30265 100 - 1 GAGCGGAGGGCACCUCCUCC------UCCUC---CUGUUACUGCUCCUUGCCUUUUGAAUGCUCCCCGCUGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGG ((((((((((....))))))------..(..---..).....))))........((((((((.....)).((((((....))))))))))))....((((....)))). ( -28.20) >DroYak_CAF1 33681 103 - 1 CAGCGGAGGGCACCUCCUCC------UCCACCAUCUCUUUCUGUUCCUUGCUUUUUGAAUGCUUCCCGCUGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGG ....((((((.......)))------))).((((((.....(((....((....((((((((.....)).((((((....))))))))))))....))..))))))))) ( -29.00) >consensus GAGCGGAGGGCACCUCCUCC______UCCUC___CUGUUUCUGCUCCUUGCCUUUUGAAUGCUCCCCGCUGGCAGCAGUCGUUGCCAUUCAAUCCUCAUCACAAGAUGG .(((((.(((((..............................((.....))(....)..))))).)))))((((((....))))))..........((((....)))). (-25.75 = -25.12 + -0.62)

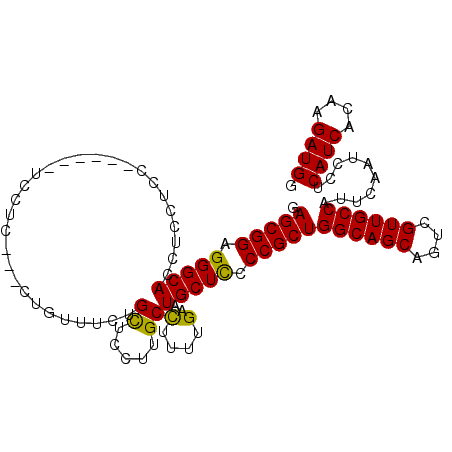
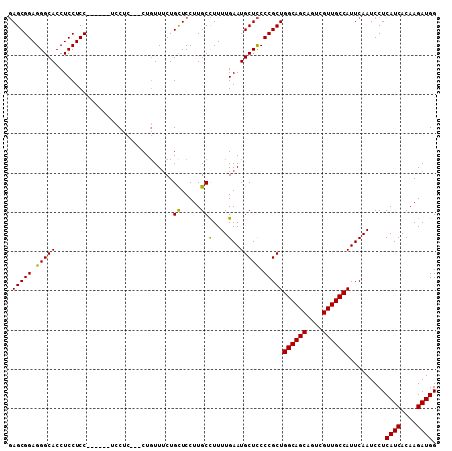
| Location | 24,921,520 – 24,921,621 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 99.01 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -31.40 |
| Energy contribution | -31.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24921520 101 + 27905053 UAUAGGCAGAUGCAGGAGGCGCUUGUUAAAUUCUUUUGCAUUUAGUUUUGCUGUCGAUAAUGAUGCUGCGUCGGCAGCAGACUUGGAAACACUGGAGCCCA ....(((((((((((((((............))))))))))))(((((.(((((((((...........))))))))))))))((....)).....))).. ( -33.10) >DroSec_CAF1 34628 101 + 1 UGUAGGCAGAUGCAGGAGGCGCUUGUUAAAUUCUUUUGCAUUUAGUUUUGCUGUCGAUAAUGAUGCUGCGUCGGCAGCAGACUUCGAAACACUGGAGCCCA ....(((((((((((((((............)))))))))))).((((((..(((........((((((....)))))))))..))))))......))).. ( -32.40) >DroSim_CAF1 30365 101 + 1 UGUAGGCAGAUGCAGGAGGCGCUUGUUAAAUUCUUUUGCAUUUAGUUUUGCUGUCGAUAAUGAUGCUGCGUCGGCAGCAGACUUCGAAACACUGGAGCCCA ....(((((((((((((((............)))))))))))).((((((..(((........((((((....)))))))))..))))))......))).. ( -32.40) >DroEre_CAF1 33358 101 + 1 UAUAGGCAGAUGCAGGAGGCGCUUGUUAAAUUCUUUUGCAUUUAGUUUUGCUGUCGAUAAUGAUGCUGCGUCGGCAGCAGACUUCGAAACACUGGAGCCCA ....(((((((((((((((............)))))))))))).((((((..(((........((((((....)))))))))..))))))......))).. ( -32.40) >DroYak_CAF1 33784 101 + 1 UAUAGGCAGAUGCAGGAGGCGCUUGUUAAAUUCUUUUGCAUUUAGUUUUGCUGUCGAUAAUGAUGCUGCGUCGGCAGCAGACUUCGAAACACUGGAGCCCA ....(((((((((((((((............)))))))))))).((((((..(((........((((((....)))))))))..))))))......))).. ( -32.40) >consensus UAUAGGCAGAUGCAGGAGGCGCUUGUUAAAUUCUUUUGCAUUUAGUUUUGCUGUCGAUAAUGAUGCUGCGUCGGCAGCAGACUUCGAAACACUGGAGCCCA ....(((((((((((((((............))))))))))))(((((.(((((((((...........)))))))))))))).............))).. (-31.40 = -31.40 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:33:58 2006