
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,918,086 – 24,918,246 |
| Length | 160 |
| Max. P | 0.893504 |

| Location | 24,918,086 – 24,918,206 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24918086 120 + 27905053 GAGAAGUUGUGGCAGCGAAAUCAGCCAUCCGCAAGCUGAAUCAGCAACAACAAUCCGAAAACAAUAGAUUUCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGAAAGGAAAAUACAAG ..((((((((((..((.......))...))))..((((...)))).....................)))))).....................((((((((..(....)...)))))))) ( -24.20) >DroSec_CAF1 31247 120 + 1 GAGAAGUUGUGGCAGCGAAAUCAGCCAUCCACAAGCUGAAUCAGCAACAACACAACGAAAACAAUGGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGAAAGGAAAAUACAAG .....((((((...((....(((((.........)))))....)).....)))))).....((....((((......))))...)).......((((((((..(....)...)))))))) ( -25.90) >DroSim_CAF1 26992 120 + 1 GAGAAGUUGUGGCAGCGAAAUCAGCCAUCCACAAGCUGAAUCAGCAACAACACAACGAAAACAAUAGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGAAAGGAAAAUACAAG .....((((((...((....(((((.........)))))....)).....)))))).....((....((((......))))...)).......((((((((..(....)...)))))))) ( -25.90) >DroEre_CAF1 29937 120 + 1 GAGAAGUUGUGGCUGCGAAAUCAGCCAUCCACAAGCUGAAUCAGCAGCAACAAUCCGAAAACAAUUGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGGAAUGAAAAUACAAG ......((((((((((....(((((.........)))))....)))))..((.((((...(((((((.((....)).)))).)))..(((((....)))))..)))).))....))))). ( -27.50) >DroYak_CAF1 30336 120 + 1 GAGAAGUUGUGGCUGCGAAAUCAGCCAUCCACAAGCUGAAUCAGCAGCAACAAUCCGAAAACAAUAGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGAUGAAAGGAAAAUACAAG ........(((((((......))))))).((((.((((......))))..(((((...........)))))...........)))).......((((((((..(....)...)))))))) ( -26.90) >consensus GAGAAGUUGUGGCAGCGAAAUCAGCCAUCCACAAGCUGAAUCAGCAACAACAAUCCGAAAACAAUAGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGAAAGGAAAAUACAAG .....(((((....((....(((((.........)))))....)).)))))..........((....((((......))))...)).......((((((((..(....)...)))))))) (-20.52 = -20.96 + 0.44)



| Location | 24,918,086 – 24,918,206 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.44 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24918086 120 - 27905053 CUUGUAUUUUCCUUUCGUCAAUACAAGGGUAUUUCACAUAUCGGUCUUGAAAUCUAUUGUUUUCGGAUUGUUGUUGCUGAUUCAGCUUGCGGAUGGCUGAUUUCGCUGCCACAACUUCUC ((((((((..(.....)..))))))))((((....(((...((((((.((((........)))))))))).))).((.((.((((((.......)))))).)).)))))).......... ( -28.10) >DroSec_CAF1 31247 120 - 1 CUUGUAUUUUCCUUUCGUCAAUACAAGGGUAUUUCACAUAUCGGUCUUGCAAUCCAUUGUUUUCGUUGUGUUGUUGCUGAUUCAGCUUGUGGAUGGCUGAUUUCGCUGCCACAACUUCUC ((((((((..(.....)..))))))))((((....(((((.(((....((((....))))..))).)))))....((.((.((((((.......)))))).)).)))))).......... ( -27.50) >DroSim_CAF1 26992 120 - 1 CUUGUAUUUUCCUUUCGUCAAUACAAGGGUAUUUCACAUAUCGGUCUUGCAAUCUAUUGUUUUCGUUGUGUUGUUGCUGAUUCAGCUUGUGGAUGGCUGAUUUCGCUGCCACAACUUCUC ((((((((..(.....)..))))))))((((....(((((.(((....((((....))))..))).)))))....((.((.((((((.......)))))).)).)))))).......... ( -27.50) >DroEre_CAF1 29937 120 - 1 CUUGUAUUUUCAUUCCGUCAAUACAAGGGUAUUUCACAUAUCGGUCUUGCAAUCAAUUGUUUUCGGAUUGUUGCUGCUGAUUCAGCUUGUGGAUGGCUGAUUUCGCAGCCACAACUUCUC ((((((((..(.....)..))))))))(((..((((((.((((((...((((.(((((.......))))))))).))))))......))))))((((((......))))))..))).... ( -32.30) >DroYak_CAF1 30336 120 - 1 CUUGUAUUUUCCUUUCAUCAAUACAAGGGUAUUUCACAUAUCGGUCUUGCAAUCUAUUGUUUUCGGAUUGUUGCUGCUGAUUCAGCUUGUGGAUGGCUGAUUUCGCAGCCACAACUUCUC ((((((((...........))))))))(((..((((((.((((((...(((((((.........)))))))....))))))......))))))((((((......))))))..))).... ( -30.70) >consensus CUUGUAUUUUCCUUUCGUCAAUACAAGGGUAUUUCACAUAUCGGUCUUGCAAUCUAUUGUUUUCGGAUUGUUGUUGCUGAUUCAGCUUGUGGAUGGCUGAUUUCGCUGCCACAACUUCUC ((((((((...........))))))))(((.........((((((...(((((....((....))....))))).))))))((((((.......)))))).......))).......... (-22.48 = -22.44 + -0.04)

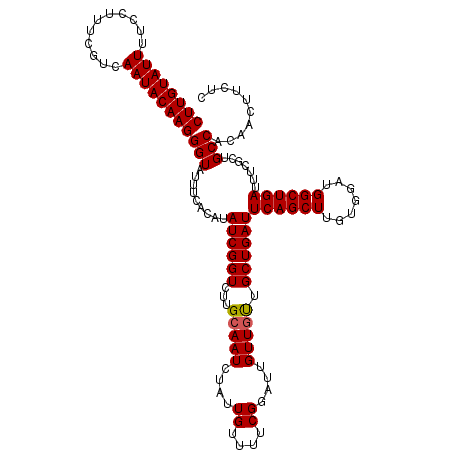

| Location | 24,918,126 – 24,918,246 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -22.48 |
| Energy contribution | -24.76 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24918126 120 + 27905053 UCAGCAACAACAAUCCGAAAACAAUAGAUUUCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGAAAGGAAAAUACAAGGUCAUUAUCGUCUAGAAAUAAUUGCGUGUGAUUUUGGGUA ............(((((((((((....(((((.((((.((((.((((.....(((((((((..(....)...))))))))))))))))))))).))))).......)))..)))))))). ( -33.60) >DroSec_CAF1 31287 120 + 1 UCAGCAACAACACAACGAAAACAAUGGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGAAAGGAAAAUACAAGGUCAUUAUCGUCUGAAAAUAAUUGCGUGUGAUUUUGGGUA ....(((...((((.((....((((..(((...((((.((((.((((.....(((((((((..(....)...)))))))))))))))))))))...))).))))))))))...))).... ( -29.40) >DroSim_CAF1 27032 120 + 1 UCAGCAACAACACAACGAAAACAAUAGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGAAAGGAAAAUACAAGGUCAUUAUCGUCUGGAAAUAAUUGCGUGUGAUUUUGGGUA ....(((...((((.((....((((..(((...((((.((((.((((.....(((((((((..(....)...)))))))))))))))))))))...))).))))))))))...))).... ( -28.20) >DroEre_CAF1 29977 120 + 1 UCAGCAGCAACAAUCCGAAAACAAUUGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGGAAUGAAAAUACAAGGCCAUUAUCGUCUGGAAAUAAUUGCGUGUGAUUUUGGGUA ............(((((((((((...(((((..((((.((((.(((......(((((((((...........))))))))).))))))))))).....)))))...)))..)))))))). ( -24.10) >DroYak_CAF1 30376 120 + 1 UCAGCAGCAACAAUCCGAAAACAAUAGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGAUGAAAGGAAAAUACAAGGUCAUUAUCGUCUGCAUAUAAUUGCGUGUGAUUUUUGGUA .......(((.((((((....((((....((((.(((.((((.((((.....(((((((((..(....)...))))))))))))))))))))))))....))))..)).)))).)))... ( -28.30) >consensus UCAGCAACAACAAUCCGAAAACAAUAGAUUGCAAGACCGAUAUGUGAAAUACCCUUGUAUUGACGAAAGGAAAAUACAAGGUCAUUAUCGUCUGGAAAUAAUUGCGUGUGAUUUUGGGUA ............(((((((((((...(((((..((((.((((.((((.....(((((((((..(....)...))))))))))))))))))))).....)))))...)))..)))))))). (-22.48 = -24.76 + 2.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:33:46 2006