
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,915,978 – 24,916,207 |
| Length | 229 |
| Max. P | 0.994071 |

| Location | 24,915,978 – 24,916,088 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -30.39 |
| Energy contribution | -30.70 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24915978 110 - 27905053 GACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUUUGCCGGC----UGCUUGUGCGGCUA------UGGCCAAUUGAAAAUGCAUUAUUCAGGUCUCAAAUGAAC ((((((...((....))...(((...(((((((((.........)))))))((((((----(((....)))))).------.))).....))...)))......)))))).......... ( -35.80) >DroSec_CAF1 29157 116 - 1 GACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUUUGCCGGC----UGCUUGUGCGGCUAUGGCUAUGGCCAAUUGAAAAUGCAUUAUUCAGGUCUCAAAUGAAC ((((((...((....))...(((..(.((((((((.........)))))))).)(((----(((....)))))).((((....))))........)))......)))))).......... ( -39.20) >DroEre_CAF1 27829 114 - 1 GACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUAUGUUGGGUUUGUACUUGUGCGGCUA------UGGCCAAUUGAAAAUACAUUAUUCAGGUCUCAAAUGAAC ((((((.........((((.(((((((((((((.((((.(((.....))).)))))))))).)))).)))(((..------..)))..))))............)))))).......... ( -34.20) >DroYak_CAF1 28193 114 - 1 GACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUAUGUUGGGUUUGUGCUUGUGCGGCUA------UGGCCAAUUGAAAAUGCAUUAUUCAGGUCUCAAAUGAAC ((((((.........((((.(((((((((((((.((((.(((.....))).)))))))))).)))).)))(((..------..)))..))))............)))))).......... ( -33.60) >consensus GACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUAUGCCGGC____UGCUUGUGCGGCUA______UGGCCAAUUGAAAAUGCAUUAUUCAGGUCUCAAAUGAAC ((((((.........((((.(((((((((((((((.........))))))))..........)))).)))(((((......)))))..))))............)))))).......... (-30.39 = -30.70 + 0.31)



| Location | 24,916,017 – 24,916,127 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -37.68 |
| Energy contribution | -38.00 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24916017 110 + 27905053 -----UAGCCGCACAAGCA----GCCGGCAAACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAACGCCC -----..((((((.(((((----(..((((((((((.........)))))))).)))))))))).)).)).((((.....(((....))).(((((((..-....)))))))....)))) ( -43.50) >DroSec_CAF1 29197 115 + 1 AGCCAUAGCCGCACAAGCA----GCCGGCAAACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAAAGCCC .(((...((((((.(((((----(..((((((((((.........)))))))).)))))))))).)).))..))).(((.(((....))).(((((((..-....)))))))....))). ( -44.70) >DroEre_CAF1 27868 114 + 1 -----UAGCCGCACAAGUACAAACCCAACAUACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAACGCCC -----..((.(((.((((.((((((((.......(((....))))).)))))))))))))(((((...((((((.....)))))).)))))(((((((..-....)))))))....)).. ( -37.01) >DroYak_CAF1 28232 115 + 1 -----UAGCCGCACAAGCACAAACCCAACAUACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCUCUUAACCACAGCAAACGCCC -----..((((((.(((((((((((((.......(((....))))).))))))....))))))).)).)).((((.....(((....))).(((((((.......)))))))....)))) ( -35.71) >consensus _____UAGCCGCACAAGCA____CCCAACAAACCGCUUAUAAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU_UUAACCACAGCAAACGCCC .......((((((.(((((.........((((((((.........))))))))....))))))).)).)).((((.....(((....))).(((((((.......)))))))....)))) (-37.68 = -38.00 + 0.31)

| Location | 24,916,017 – 24,916,127 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -46.38 |
| Consensus MFE | -40.60 |
| Energy contribution | -41.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24916017 110 - 27905053 GGGCGUUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUUUGCCGGC----UGCUUGUGCGGCUA----- ((((....((((((((...-.))))))))((((....))))....))))((....))........(.((((((((.........)))))))).)(((----(((....)))))).----- ( -46.70) >DroSec_CAF1 29197 115 - 1 GGGCUUUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUUUGCCGGC----UGCUUGUGCGGCUAUGGCU ((((....((((((((...-.))))))))((((....))))....))))((((.((....))..))))(((((((.........)))))))((((((----(((....))))))..))). ( -49.60) >DroEre_CAF1 27868 114 - 1 GGGCGUUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUAUGUUGGGUUUGUACUUGUGCGGCUA----- ((((....((((((((...-.))))))))((((....))))....)))).....(((...(((((((((((((.((((.(((.....))).)))))))))).)))).))))))..----- ( -44.70) >DroYak_CAF1 28232 115 - 1 GGGCGUUUGCUGUGGUUAAGAGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUAUGUUGGGUUUGUGCUUGUGCGGCUA----- ((((....((((((((.....))))))))((((....))))....))))...((((..(((((....((((((.((((.(((.....))).)))))))))))))))..))))...----- ( -44.50) >consensus GGGCGUUUGCUGUGGUUAA_AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUUAUAAGCGGUAUGCCGGC____UGCUUGUGCGGCUA_____ ((((....((((((((.....))))))))((((....))))....))))...((((..(((((..(.((((((((.........)))))))).).......)))))..))))........ (-40.60 = -41.10 + 0.50)


| Location | 24,916,048 – 24,916,167 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -45.15 |
| Consensus MFE | -43.41 |
| Energy contribution | -43.73 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24916048 119 + 27905053 AAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAACGCCCGCCAAACCACGACAACCACACAAAGUGGUGGCAAAGCCAG ..(((((((((((.....((((....))))(((((.....(((....))).(((((((..-....)))))))....))))).))))))......(((((.....))))).))..)))... ( -43.50) >DroSec_CAF1 29233 119 + 1 AAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAAAGCCCGCCAAACCACGACAACCACACAAAGUGGUGGCAAAGCCAG ..(((((((((((.(...(((((...((((((((.....)))))).))...(((((((..-....))))))).)))))..).))))))......(((((.....))))).))..)))... ( -43.60) >DroEre_CAF1 27903 115 + 1 AAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAACGCCCGCCAAACCACGGCAACCACACAAAGUGGUGGCAAAG---- ..((((.((((((.....((((....))))(((((.....(((....))).(((((((..-....)))))))....))))).)))))).)))).(((((.....))))).......---- ( -47.00) >DroYak_CAF1 28267 116 + 1 AAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCUCUUAACCACAGCAAACGCCCGCCAAGCCACGGCAACCACACAAAGUGGUGGCAGAG---- ..((((.((((((.....((((....))))(((((.....(((....))).(((((((.......)))))))....))))).)))))).)))).(((((.....))))).......---- ( -46.50) >consensus AAGCUGCGGUUUGACUUUGCUUUGACGAGUCGGGCAGGUCCUGACAUCAGAGCUGUGGCU_UUAACCACAGCAAACGCCCGCCAAACCACGACAACCACACAAAGUGGUGGCAAAG____ ..((((.((((((.....((((....))))(((((.....(((....))).(((((((.......)))))))....))))).)))))).)))).(((((.....)))))........... (-43.41 = -43.73 + 0.31)


| Location | 24,916,048 – 24,916,167 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -51.15 |
| Consensus MFE | -49.95 |
| Energy contribution | -49.95 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24916048 119 - 27905053 CUGGCUUUGCCACCACUUUGUGUGGUUGUCGUGGUUUGGCGGGCGUUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUU .(((((((((.(((((.....)))))((.((.((((.((((((.....((((((((...-.))))))))((((....)))).)))))).)))))).))..)))))))))........... ( -52.60) >DroSec_CAF1 29233 119 - 1 CUGGCUUUGCCACCACUUUGUGUGGUUGUCGUGGUUUGGCGGGCUUUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUU .(((((((((.(((((.....)))))((.((.((((.((((((.....((((((((...-.))))))))((((....)))).)))))).)))))).))..)))))))))........... ( -52.60) >DroEre_CAF1 27903 115 - 1 ----CUUUGCCACCACUUUGUGUGGUUGCCGUGGUUUGGCGGGCGUUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUU ----....((.(((((.....))))).)).((((((((((((((....((((((((...-.))))))))((((....))))....))))((....))........))))))))))..... ( -51.80) >DroYak_CAF1 28267 116 - 1 ----CUCUGCCACCACUUUGUGUGGUUGCCGUGGCUUGGCGGGCGUUUGCUGUGGUUAAGAGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUU ----....((.(((((.....))))).)).((((.(((((((((....((((((((.....))))))))((((....))))....))))((....))........))))).))))..... ( -47.60) >consensus ____CUUUGCCACCACUUUGUGUGGUUGCCGUGGUUUGGCGGGCGUUUGCUGUGGUUAA_AGCCACAGCUCUGAUGUCAGGACCUGCCCGACUCGUCAAAGCAAAGUCAAACCGCAGCUU ........((.(((((.....))))).)).((((((((((((((....((((((((.....))))))))((((....))))....))))((....))........))))))))))..... (-49.95 = -49.95 + -0.00)

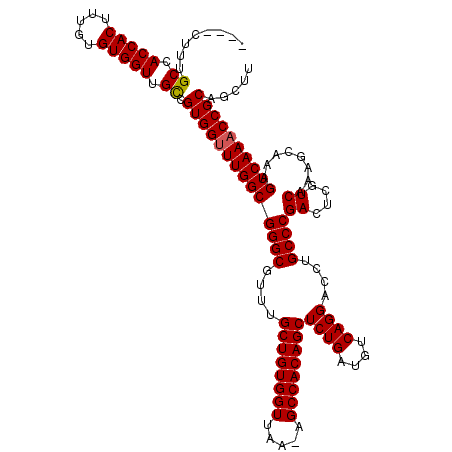
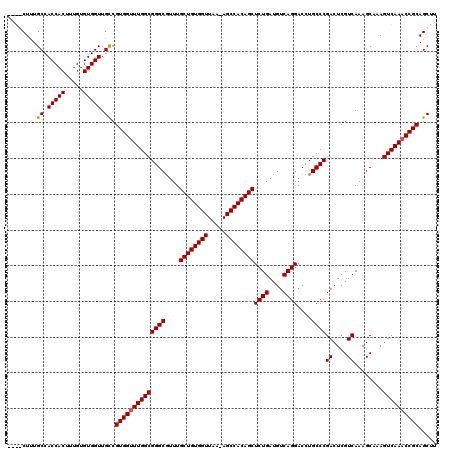
| Location | 24,916,088 – 24,916,207 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -36.06 |
| Energy contribution | -36.00 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24916088 119 + 27905053 CUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAACGCCCGCCAAACCACGACAACCACACAAAGUGGUGGCAAAGCCAGCCAACGAUUCCGCAAGAUGUGUUAAUGAUUUUGGCGGAUA (((....))).(((((((..-....)))))))......((((((((...((...(((((.....)))))(((.......)))..))....(....).............))))))))... ( -40.10) >DroSec_CAF1 29273 119 + 1 CUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAAAGCCCGCCAAACCACGACAACCACACAAAGUGGUGGCAAAGCCAGCCAACGAUUCCGCAAGAUGUGUUAAUGAUUUUGGCGGAUA (((....))).(((((((..-....)))))))......((((((((...((...(((((.....)))))(((.......)))..))....(....).............))))))))... ( -40.10) >DroEre_CAF1 27943 115 + 1 CUGACAUCAGAGCUGUGGCU-UUAACCACAGCAAACGCCCGCCAAACCACGGCAACCACACAAAGUGGUGGCAAAG----CCAACGAUUCCACAAGAUGUGUUAAUGAUUUUGGCGGAUA (((....))).(((((((..-....)))))))......((((((((....(((.(((((.....)))))......)----))..((.(..(((.....)))..).))..))))))))... ( -39.80) >DroYak_CAF1 28307 116 + 1 CUGACAUCAGAGCUGUGGCUCUUAACCACAGCAAACGCCCGCCAAGCCACGGCAACCACACAAAGUGGUGGCAGAG----CCAACGAUUCCGUAAGAUGUGUUAAUGAUUUUGGCGGAUA (((....))).(((((((.......)))))))......(((((((((((((....)(((.....))))))))....----..((((....(....)...)))).......)))))))... ( -40.30) >consensus CUGACAUCAGAGCUGUGGCU_UUAACCACAGCAAACGCCCGCCAAACCACGACAACCACACAAAGUGGUGGCAAAG____CCAACGAUUCCGCAAGAUGUGUUAAUGAUUUUGGCGGAUA (((....))).(((((((.......)))))))......((((((((...((...(((((.....)))))(((.......)))..))....(((.....)))........))))))))... (-36.06 = -36.00 + -0.06)


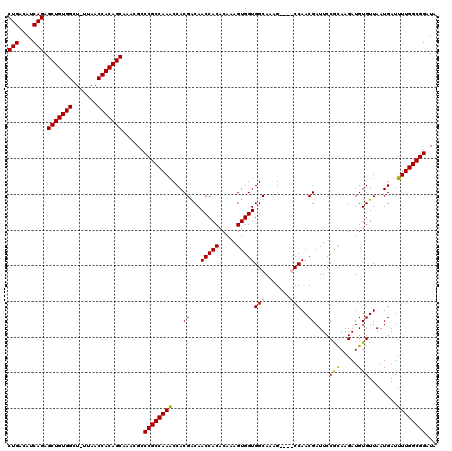
| Location | 24,916,088 – 24,916,207 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -44.35 |
| Consensus MFE | -40.33 |
| Energy contribution | -40.58 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24916088 119 - 27905053 UAUCCGCCAAAAUCAUUAACACAUCUUGCGGAAUCGUUGGCUGGCUUUGCCACCACUUUGUGUGGUUGUCGUGGUUUGGCGGGCGUUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAG ....(((((((.((((((((((((...(((....)))(((.((((...)))))))....)))).))))..)))))))))))((((((.((((((((...-.))))))))...)))))).. ( -44.60) >DroSec_CAF1 29273 119 - 1 UAUCCGCCAAAAUCAUUAACACAUCUUGCGGAAUCGUUGGCUGGCUUUGCCACCACUUUGUGUGGUUGUCGUGGUUUGGCGGGCUUUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAG ..(((((((((.((((((((((((...(((....)))(((.((((...)))))))....)))).))))..))))))))))))).....((((((((...-.)))))))).(((....))) ( -42.30) >DroEre_CAF1 27943 115 - 1 UAUCCGCCAAAAUCAUUAACACAUCUUGUGGAAUCGUUGG----CUUUGCCACCACUUUGUGUGGUUGCCGUGGUUUGGCGGGCGUUUGCUGUGGUUAA-AGCCACAGCUCUGAUGUCAG ....(((((((.((((...(((.....)))........((----(...((((((.....).))))).))))))))))))))((((((.((((((((...-.))))))))...)))))).. ( -47.50) >DroYak_CAF1 28307 116 - 1 UAUCCGCCAAAAUCAUUAACACAUCUUACGGAAUCGUUGG----CUCUGCCACCACUUUGUGUGGUUGCCGUGGCUUGGCGGGCGUUUGCUGUGGUUAAGAGCCACAGCUCUGAUGUCAG ....((((((......((((...((.....))...))))(----(((.((.(((((.....))))).)).).)))))))))((((((.((((((((.....))))))))...)))))).. ( -43.00) >consensus UAUCCGCCAAAAUCAUUAACACAUCUUGCGGAAUCGUUGG____CUUUGCCACCACUUUGUGUGGUUGCCGUGGUUUGGCGGGCGUUUGCUGUGGUUAA_AGCCACAGCUCUGAUGUCAG ....(((((((.((((((((...((.....))...)))).........((.(((((.....))))).)).)))))))))))((((((.((((((((.....))))))))...)))))).. (-40.33 = -40.58 + 0.25)

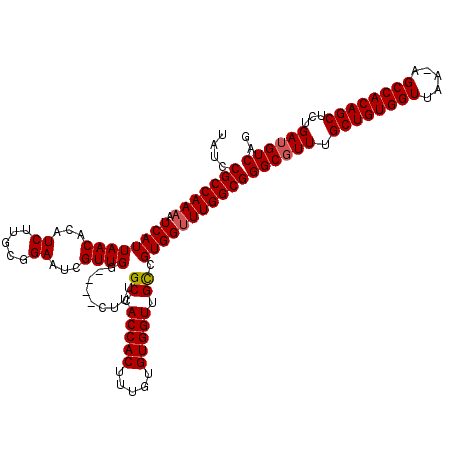

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:33:39 2006