| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
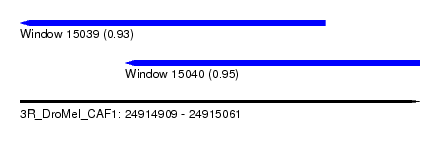
| Location | 24,914,909 – 24,915,061 |
| Length | 152 |
| Max. P | 0.952306 |

| Location | 24,914,909 – 24,915,025 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -20.88 |
| Consensus MFE | -17.20 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24914909 116 - 27905053 UCAAGACGUGCGCUUUACACUUCUGCGCCCCAA-AAUGCCCCAACAAUCACAACAUACAGAAGAAGAAAAAAAA---CUACACAUUGAAAUUACAAACUGAAACGCGCAGGUCUAAUAGU ...((((.(((((......((((((........-.......................))))))...........---.......(((......)))........))))).))))...... ( -20.28) >DroSec_CAF1 28070 119 - 1 UCAAGACGUGCGCUUUACACUUCUGCGCCCCAA-AAUGCCCCAACAAUAACAACAUUCAAAAGAAGAAAAAAAAACGCUACAGUUUGAAAUUACAAACCGAAACGCGCAGGUCUAAUAGU ...((((.(((((......(((((.........-...........................)))))..............(.(((((......))))).)....))))).))))...... ( -22.02) >DroEre_CAF1 26714 114 - 1 UCAAGACGUGCGCUUUACACUUCUGCGCCCCAAAAAUGCCCCAACAAUAACAACAUACAGAAAAAAAAA----CACGCUAC--AGUGAAAUUACAAACUGAAACGCGCAGGUCUAAUAAU ...((((.(((((.......(((((...........((......))...........))))).......----.......(--(((..........))))....))))).))))...... ( -20.95) >DroYak_CAF1 27042 119 - 1 UCAAGACGUGCGCUUUACACUUCUGCGCCCCAA-AAUGCCCCAACAAUAACAACAUACAGAAGAAAAAAAAACCCCGCUACACAUUGAAAUUACAAACCGAAACGCGCAGGUCUAAUAGU ...((((.(((((......((((((........-.......................)))))).....................(((......)))........))))).))))...... ( -20.28) >consensus UCAAGACGUGCGCUUUACACUUCUGCGCCCCAA_AAUGCCCCAACAAUAACAACAUACAGAAGAAAAAAAAAAAACGCUACACAUUGAAAUUACAAACCGAAACGCGCAGGUCUAAUAGU ...((((.(((((......((((((...........((......))...........)))))).....................(((......)))........))))).))))...... (-17.20 = -17.95 + 0.75)



| Location | 24,914,949 – 24,915,061 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -23.05 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24914949 112 - 27905053 UAUCUGUGUGUUUUGUUAUAUUGACGUAUCUGAGCCUCAAGACGUGCGCUUUACACUUCUGCGCCCCAA-AAUGCCCCAACAAUCACAACAUACAGAAGAAGAAAAAAAA---CUA ..(((((((((((((((......((((...((.....))..))))((((...........)))).....-........)))))....)))))))))).............---... ( -23.20) >DroSec_CAF1 28110 115 - 1 UAUCUGAGUGUUUUGUUAUAUUGACGUAUCUGAGCCUCAAGACGUGCGCUUUACACUUCUGCGCCCCAA-AAUGCCCCAACAAUAACAACAUUCAAAAGAAGAAAAAAAAACGCUA ....(((((((..((((((.(((((((...((.....))..))))((((...........)))).....-.......)))..)))))))))))))..................... ( -20.00) >DroEre_CAF1 26752 112 - 1 UAUCUGUGUGUUUUGUUAUAUUGACGUAUCUGAGCCUCAAGACGUGCGCUUUACACUUCUGCGCCCCAAAAAUGCCCCAACAAUAACAACAUACAGAAAAAAAAA----CACGCUA ..(((((((((..((((((.(((((((...((.....))..))))((((...........)))).............)))..)))))))))))))))........----....... ( -24.90) >DroYak_CAF1 27082 115 - 1 UAUCUGUGUGUUUUGUUAUAUUGACGUAUCUGAGCCUCAAGACGUGCGCUUUACACUUCUGCGCCCCAA-AAUGCCCCAACAAUAACAACAUACAGAAGAAAAAAAAACCCCGCUA ..(((((((((..((((((.(((((((...((.....))..))))((((...........)))).....-.......)))..)))))))))))))))................... ( -24.90) >consensus UAUCUGUGUGUUUUGUUAUAUUGACGUAUCUGAGCCUCAAGACGUGCGCUUUACACUUCUGCGCCCCAA_AAUGCCCCAACAAUAACAACAUACAGAAGAAAAAAAAAAAACGCUA ..(((((((((..((((((.(((((((...((.....))..))))((((...........)))).............)))..)))))))))))))))................... (-23.05 = -23.18 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:33:32 2006