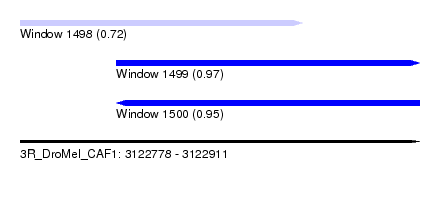
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,122,778 – 3,122,911 |
| Length | 133 |
| Max. P | 0.967954 |

| Location | 3,122,778 – 3,122,872 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 90.81 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -28.09 |
| Energy contribution | -28.65 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
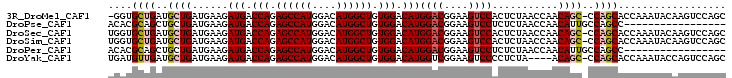
>3R_DroMel_CAF1 3122778 94 + 27905053 GACACACACACACACGCACAGACGCAGCUGCU------GGUGCUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUC ................(((((...((((....------...))))...))).))...(((.(((.((((((....)))))).))).))).(((....))) ( -30.50) >DroSec_CAF1 10382 94 + 1 GACACAC------ACGCACAGACGCAGCUGCUGGUGCUGGUGCUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUC ....(((------(.((((((((.((((.......)))))).))).))))).))...(((.(((.((((((....)))))).))).))).(((....))) ( -35.20) >DroSim_CAF1 15689 94 + 1 GACACAC------ACGCACAGACGCAGCUGCUGGUGCUGGUGCUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUC ....(((------(.((((((((.((((.......)))))).))).))))).))...(((.(((.((((((....)))))).))).))).(((....))) ( -35.20) >DroYak_CAF1 14069 88 + 1 GACACAC------ACGCACAGACGCAGCUGC------UGAUGUUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGUCGGAAGUC (((...(------(.(((..(((((((...)------)).))))..))))).(((..(((.(((.((((((....)))))).))).)))..)))...))) ( -29.30) >consensus GACACAC______ACGCACAGACGCAGCUGCU_____UGGUGCUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUC ................(((((...((((.(((......)))))))...))).))...(((.(((.((((((....)))))).))).))).(((....))) (-28.09 = -28.65 + 0.56)

| Location | 3,122,810 – 3,122,911 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -31.41 |
| Energy contribution | -31.88 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3122810 101 + 27905053 -GGUGCUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUCCACUCUAACCAACAGC-CCAGCACCAAAUACAAGUCCAGC -(((((((..((((.((...(((.(((.((((((....)))))).))).)))((((....))))........)).))))-.)))))))............... ( -45.70) >DroPse_CAF1 10245 86 + 1 ACACGCAGCUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUCCUCUCUAACCAACAUUGCCAGCC----------------- .......((((..((((...(((.(((.((((((....)))))).))).)))((((....))))...........))))..)))).----------------- ( -33.90) >DroSec_CAF1 10413 102 + 1 UGGUGCUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUCCACUCUAACCAACAGC-CCAGCACCAAAUACAAGUCCAGC ((((((((..((((.((...(((.(((.((((((....)))))).))).)))((((....))))........)).))))-.)))))))).............. ( -46.40) >DroSim_CAF1 15720 102 + 1 UGGUGCUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUCCACUCUAACCAACAGC-CCAGCACCAAAUACAAGUCCAGC ((((((((..((((.((...(((.(((.((((((....)))))).))).)))((((....))))........)).))))-.)))))))).............. ( -46.40) >DroPer_CAF1 10208 86 + 1 ACACGCAGCUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUCCUCUCUAACCAACAUUGCCAGCC----------------- .......((((..((((...(((.(((.((((((....)))))).))).)))((((....))))...........))))..)))).----------------- ( -33.90) >DroYak_CAF1 14094 98 + 1 UGAUGUUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGUCGGAAGUCCCCUCUA----ACAGC-CCAGCACCAAAUACCAGUCCAGC ((.(((((..((((.((.(((((.(((.((((((....)))))).))).)))((.(....).)).)).))----.))))-.))))).)).............. ( -31.90) >consensus UGAUGCUGAUGCUGAUGAAGAUGACCAGAGCCAUGGACAUGGCUGUGGACAUGGACGGAAGUCCACUCUAACCAACAGC_CCAGCACCAAAUACAAGUCCAGC ....((((..((((......(((.(((.((((((....)))))).))).)))((((....))))...........))))..)))).................. (-31.41 = -31.88 + 0.47)
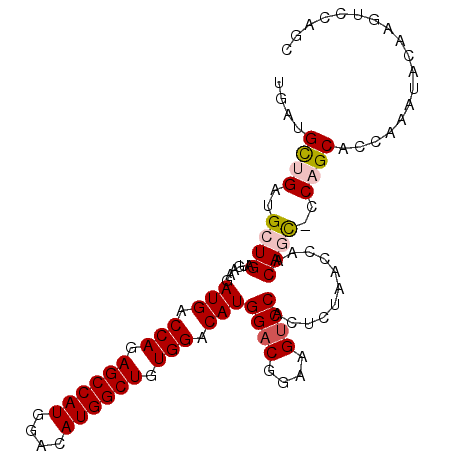
| Location | 3,122,810 – 3,122,911 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -30.87 |
| Energy contribution | -33.95 |
| Covariance contribution | 3.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3122810 101 - 27905053 GCUGGACUUGUAUUUGGUGCUGG-GCUGUUGGUUAGAGUGGACUUCCGUCCAUGUCCACAGCCAUGUCCAUGGCUCUGGUCAUCUUCAUCAGCAUCAGCACC- ...............((((((((-((((.(((..(((((((((....))))))(.(((.((((((....)))))).))).).)))))).)))).))))))))- ( -43.30) >DroPse_CAF1 10245 86 - 1 -----------------GGCUGGCAAUGUUGGUUAGAGAGGACUUCCGUCCAUGUCCACAGCCAUGUCCAUGGCUCUGGUCAUCUUCAUCAGCAGCUGCGUGU -----------------.....(((.(((((((..((((((((....)))).((.(((.((((((....)))))).))).)))))).)))))))..))).... ( -33.20) >DroSec_CAF1 10413 102 - 1 GCUGGACUUGUAUUUGGUGCUGG-GCUGUUGGUUAGAGUGGACUUCCGUCCAUGUCCACAGCCAUGUCCAUGGCUCUGGUCAUCUUCAUCAGCAUCAGCACCA ..............(((((((((-((((.(((..(((((((((....))))))(.(((.((((((....)))))).))).).)))))).)))).))))))))) ( -45.10) >DroSim_CAF1 15720 102 - 1 GCUGGACUUGUAUUUGGUGCUGG-GCUGUUGGUUAGAGUGGACUUCCGUCCAUGUCCACAGCCAUGUCCAUGGCUCUGGUCAUCUUCAUCAGCAUCAGCACCA ..............(((((((((-((((.(((..(((((((((....))))))(.(((.((((((....)))))).))).).)))))).)))).))))))))) ( -45.10) >DroPer_CAF1 10208 86 - 1 -----------------GGCUGGCAAUGUUGGUUAGAGAGGACUUCCGUCCAUGUCCACAGCCAUGUCCAUGGCUCUGGUCAUCUUCAUCAGCAGCUGCGUGU -----------------.....(((.(((((((..((((((((....)))).((.(((.((((((....)))))).))).)))))).)))))))..))).... ( -33.20) >DroYak_CAF1 14094 98 - 1 GCUGGACUGGUAUUUGGUGCUGG-GCUGU----UAGAGGGGACUUCCGACCAUGUCCACAGCCAUGUCCAUGGCUCUGGUCAUCUUCAUCAGCAUCAACAUCA ........(((..((((((((((-.....----..(((....)))..((..(((.(((.((((((....)))))).))).)))..)).)))))))))).))). ( -34.20) >consensus GCUGGACUUGUAUUUGGUGCUGG_GCUGUUGGUUAGAGUGGACUUCCGUCCAUGUCCACAGCCAUGUCCAUGGCUCUGGUCAUCUUCAUCAGCAUCAGCACCA ...............((((((((.((((.(((..(((..((((....)))).((.(((.((((((....)))))).))).)))))))).)))).)))))))). (-30.87 = -33.95 + 3.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:04 2006