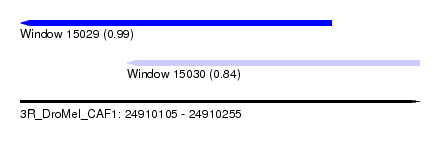
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,910,105 – 24,910,255 |
| Length | 150 |
| Max. P | 0.994005 |

| Location | 24,910,105 – 24,910,222 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.80 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -33.94 |
| Energy contribution | -34.18 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24910105 117 - 27905053 AUCGCCUUGCCACCU---CCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGAUAAUUAGUUUGCUUUCGGUGGCAGCUCCCAGAGCAUCCGA .(((...(((((((.---....((((((....((......(((((..(((((.((((........)))).)))))))))).....)).))))))..)))))))((((...))))...))) ( -36.20) >DroSec_CAF1 21700 120 - 1 AUCGCGUUGCCACCUCCUCCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGAUAAUUAGUUUGCCUUCGGUGGCAGCUCCCAGAGCAUCCGA .(((...(((((((........((((((....((......(((((..(((((.((((........)))).)))))))))).....)).))))))..)))))))((((...))))...))) ( -37.10) >DroSim_CAF1 21352 120 - 1 AUCGCGUUGCCACCUCCUCCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGAUAAUUAGUUUGCCUUCGGUGGCAGCUCCCAGAGCAUCCGA .(((...(((((((........((((((....((......(((((..(((((.((((........)))).)))))))))).....)).))))))..)))))))((((...))))...))) ( -37.10) >DroEre_CAF1 21790 117 - 1 ACCACGGUGCCACCU---CCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGAUAAUUAGUUUGCCUUCAGUGGCAGCUCCCAGAGCAUCCGA ....(((((((((..---....((((((....((......(((((..(((((.((((........)))).)))))))))).....)).))))))...))))))((((...))))..))). ( -38.10) >DroYak_CAF1 22077 117 - 1 ACCACGGUGCCACCU---CCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGAUAAUUAGUUUGCCUUCAGUGGCAGCUCCCAGAGCAUCCGA ....(((((((((..---....((((((....((......(((((..(((((.((((........)))).)))))))))).....)).))))))...))))))((((...))))..))). ( -38.10) >consensus AUCGCGUUGCCACCU___CCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGAUAAUUAGUUUGCCUUCGGUGGCAGCUCCCAGAGCAUCCGA .......(((((((........((((((....((......(((((..(((((.((((........)))).)))))))))).....)).))))))..)))))))((((...))))...... (-33.94 = -34.18 + 0.24)


| Location | 24,910,145 – 24,910,255 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24910145 110 - 27905053 AUUUCGAUUCAGAUUCGGAUUCCC-------ACUUCCACCAUCGCCUUGCCACCU---CCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGA .....((((((((...(((.....-------...))).........(((((....---.....))))).....))))))))((((..(((((.((((........)))).))))))))). ( -26.30) >DroSec_CAF1 21740 113 - 1 AUUUCGAUGCAGAUUCGGAUUCCC-------ACUUCCACCAUCGCGUUGCCACCUCCUCCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGA ..(((((.((.(((..(((.....-------...)))...)))))((((((............)))))).....))))).(((((..(((((.((((........)))).)))))))))) ( -27.00) >DroSim_CAF1 21392 113 - 1 AUUUCGAUUCAGAUUCGGAUUCCC-------ACUUCCACCAUCGCGUUGCCACCUCCUCCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGA .....((((((((...(((.....-------...)))......(.((((((............)))))).)..))))))))((((..(((((.((((........)))).))))))))). ( -27.20) >DroEre_CAF1 21830 117 - 1 AUUUCGAUUCAGAUUCGGAUUCCCACUCUCCACUUCCACCACCACGGUGCCACCU---CCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGA .....((((((((...(((((.((............((((.....))))......---.....)).)))))..))))))))((((..(((((.((((........)))).))))))))). ( -29.00) >DroYak_CAF1 22117 110 - 1 AUUUCGAUUCAGAUUCGGAUUCCC-------ACUUCCACCACCACGGUGCCACCU---CCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGA .....((((((((...(((((...-------......(((.....)))(((....---.....))))))))..))))))))((((..(((((.((((........)))).))))))))). ( -27.90) >consensus AUUUCGAUUCAGAUUCGGAUUCCC_______ACUUCCACCAUCGCGUUGCCACCU___CCCCAGGCAAUCCCCUUUGAAUUCCCCGAGCUGUCUGUUUGUCACUCAACAUAUGGCGGGGA .....((((((((...(((...............)))..........((((............))))......))))))))((((..(((((.((((........)))).))))))))). (-23.20 = -23.40 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:33:23 2006