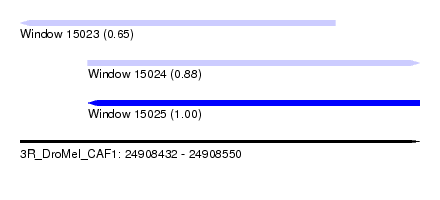
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,908,432 – 24,908,550 |
| Length | 118 |
| Max. P | 0.999539 |

| Location | 24,908,432 – 24,908,525 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 87.15 |
| Mean single sequence MFE | -16.32 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.21 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24908432 93 - 27905053 GUCCCAGCACUUCCCAUCAGUGCUGUCACUUUGGCCAUAAUUUACAUACCAAAUUUAAUUUUGAGCUUGAGAAAGAUAACAAAUAAUAUUUGC-- (((.(((((((.......)))))))...(((.(((((.((((..............)))).)).))).)))...)))................-- ( -15.24) >DroSim_CAF1 19720 93 - 1 GUCCCAGCACUUCCCAUCAGUGCUGUCACUUUGGCCAUAAUUUACAUACCAAAUUUAAUUUUGAGCUUCAGAAAGAUAACGAAUAGUAUUUGC-- ((..(((((((.......)))))))..))(((((..............))))).....(((((.....)))))....................-- ( -14.34) >DroEre_CAF1 20141 95 - 1 GUCCCAGCACUUCUCACCAGUGCUGUCACUUUGGUCAUAUUUUACAUACCAAAUUUAAUUUUAAGCUUUACAAAGGCAGCAAAUAAUAUUUGCCA ((..(((((((.......)))))))..))((((((............))))))...........((((.....)))).((((((...)))))).. ( -19.50) >DroYak_CAF1 20417 95 - 1 GUCCCAGCACUUCUCAUCAGUGCUGUCAUUUUGACCAUAAUUUACAUACCAAAUUUAAUUUUGAGCUUCACACGGACAGCAAACCAUAUUUCCAC (((((((((((.......)))))))........................((((......))))..........)))).................. ( -16.20) >consensus GUCCCAGCACUUCCCAUCAGUGCUGUCACUUUGGCCAUAAUUUACAUACCAAAUUUAAUUUUGAGCUUCACAAAGACAACAAAUAAUAUUUGC__ (((.(((((((.......)))))))....(((((..............))))).....................))).................. (-11.40 = -11.21 + -0.19)

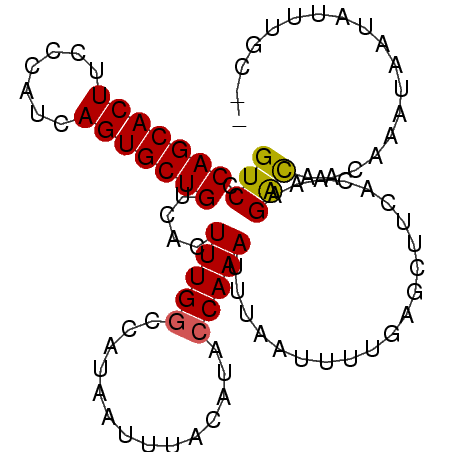

| Location | 24,908,452 – 24,908,550 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.36 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24908452 98 + 27905053 -------UUCUCAAGCUCAAAAUUAAAUUUGGUAUGUAAAUUAUGGCCAAAGUGACAGCACUGAUGGGAAGUGCUGGGACAGCAAUUUGUUAAGCCGCUUCAAUG -------.....(((((((((......)))))............(((....((..(((((((.......)))))))..))(((.....)))..)))))))..... ( -22.40) >DroSec_CAF1 20090 105 + 1 UAUAUUAUUCUCAAGCUCAAAAUUAAAUUUGGUAUGUAAAUUAUGGCCAAAGUGACAGCACUGAUGGGAAGUGCUGGGACAGCAAUUUGUUAAGCCGCUUCAAUG ............(((((((((......)))))............(((....((..(((((((.......)))))))..))(((.....)))..)))))))..... ( -22.40) >DroSim_CAF1 19740 98 + 1 -------UUCUGAAGCUCAAAAUUAAAUUUGGUAUGUAAAUUAUGGCCAAAGUGACAGCACUGAUGGGAAGUGCUGGGACAGCAAUUUGUUAAGCCGCUUCAAUG -------...(((((((((((......)))))............(((....((..(((((((.......)))))))..))(((.....)))..)))))))))... ( -27.40) >DroEre_CAF1 20163 98 + 1 -------UUGUAAAGCUUAAAAUUAAAUUUGGUAUGUAAAAUAUGACCAAAGUGACAGCACUGGUGAGAAGUGCUGGGACAGCAAUUUGUUAAGCCGCUUCAAUG -------.......((((((.......((((((((((...)))).))))))((..(((((((.......)))))))..)).........)))))).......... ( -24.40) >DroYak_CAF1 20439 98 + 1 -------GUGUGAAGCUCAAAAUUAAAUUUGGUAUGUAAAUUAUGGUCAAAAUGACAGCACUGAUGAGAAGUGCUGGGACAGCAAUUUGUUAAGCCGCUUCAAUG -------...(((((((((((......)))))............(((........(((((((.......))))))).(((((....)))))..)))))))))... ( -24.30) >consensus _______UUCUCAAGCUCAAAAUUAAAUUUGGUAUGUAAAUUAUGGCCAAAGUGACAGCACUGAUGGGAAGUGCUGGGACAGCAAUUUGUUAAGCCGCUUCAAUG ............((((...........(((((((((.....))).))))))....(((((((.......)))))))((..(((.....)))...))))))..... (-20.68 = -20.36 + -0.32)

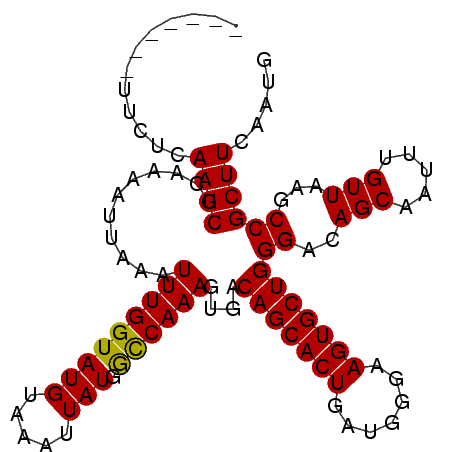
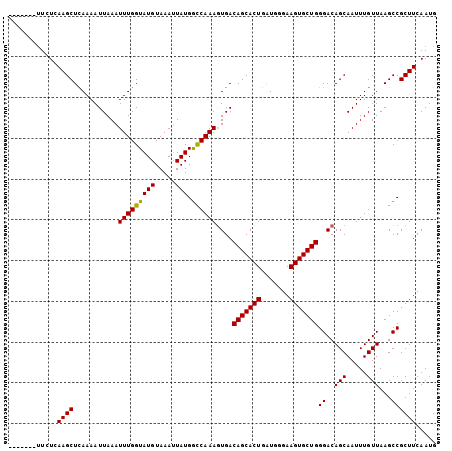
| Location | 24,908,452 – 24,908,550 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.66 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
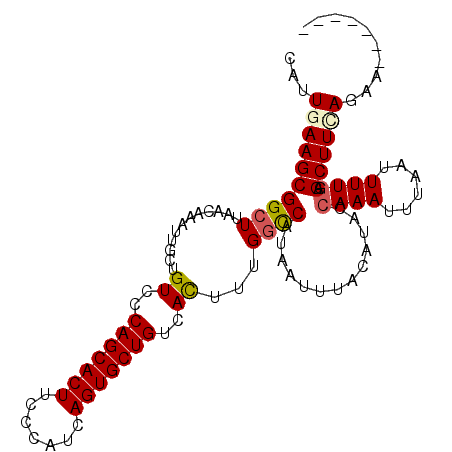
>3R_DroMel_CAF1 24908452 98 - 27905053 CAUUGAAGCGGCUUAACAAAUUGCUGUCCCAGCACUUCCCAUCAGUGCUGUCACUUUGGCCAUAAUUUACAUACCAAAUUUAAUUUUGAGCUUGAGAA------- ..((.((((((((............((..(((((((.......)))))))..))...)))).............((((......)))).)))).))..------- ( -21.36) >DroSec_CAF1 20090 105 - 1 CAUUGAAGCGGCUUAACAAAUUGCUGUCCCAGCACUUCCCAUCAGUGCUGUCACUUUGGCCAUAAUUUACAUACCAAAUUUAAUUUUGAGCUUGAGAAUAAUAUA ..((.((((((((............((..(((((((.......)))))))..))...)))).............((((......)))).)))).))......... ( -21.36) >DroSim_CAF1 19740 98 - 1 CAUUGAAGCGGCUUAACAAAUUGCUGUCCCAGCACUUCCCAUCAGUGCUGUCACUUUGGCCAUAAUUUACAUACCAAAUUUAAUUUUGAGCUUCAGAA------- ..(((((((((((............((..(((((((.......)))))))..))...)))).............((((......)))).)))))))..------- ( -25.46) >DroEre_CAF1 20163 98 - 1 CAUUGAAGCGGCUUAACAAAUUGCUGUCCCAGCACUUCUCACCAGUGCUGUCACUUUGGUCAUAUUUUACAUACCAAAUUUAAUUUUAAGCUUUACAA------- ...(((((((((..........)))((..(((((((.......)))))))..))((((((............))))))...........))))))...------- ( -20.90) >DroYak_CAF1 20439 98 - 1 CAUUGAAGCGGCUUAACAAAUUGCUGUCCCAGCACUUCUCAUCAGUGCUGUCAUUUUGACCAUAAUUUACAUACCAAAUUUAAUUUUGAGCUUCACAC------- ...((((((........((((((..(((.(((((((.......))))))).......)))..))))))......((((......)))).))))))...------- ( -20.40) >consensus CAUUGAAGCGGCUUAACAAAUUGCUGUCCCAGCACUUCCCAUCAGUGCUGUCACUUUGGCCAUAAUUUACAUACCAAAUUUAAUUUUGAGCUUCAGAA_______ ...((((((((((............((..(((((((.......)))))))..))...)))).............((((......)))).)))))).......... (-19.26 = -19.66 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:33:18 2006